Abstract
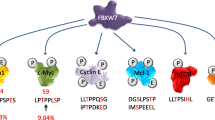
Fbw7 is a member of F-box family proteins, which constitute one subunit of Skp1, Cul1, and F-box protein (SCF) ubiquitin ligase complex. SCFFbw7 targets a set of well-known oncoproteins, including c-Myc, cyclin E, Notch, c-Jun, and Mcl-1, for ubiquitylation and degradation. Fbw7 provides specificity of the ubiquitylation of these substrate proteins via recognition of a consensus phosphorylated degron. Through regulation of several important proteins, Fbw7 controls diverse cellular processes, including cell-cycle progression, cell proliferation, differentiation, DNA damage response, maintenance of genomic stability, and neural cell stemness. As reduced Fbw7 expression level and loss-of-function mutations are found in a wide range of human cancers, Fbw7 is generally considered as a tumor suppressor. However, the exact mechanisms underlying Fbw7-induced tumor suppression is unclear. This review focuses on regulation network, biological functions, and genetic alteration of Fbw7 in connection with its role in cancer development.

Similar content being viewed by others
References
Crusio, K. M., King, B., Reavie, L. B., & Aifantis, I. (2010). The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene, 29, 4865–4873.
Hershko, A. (1983). Ubiquitin: roles in protein modification and breakdown. Cell, 34, 11–12.
Schwartz, A. L., & Ciechanover, A. (2009). Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annual Review of Pharmacology and Toxicology, 49, 73–96.
Welcker, M., & Clurman, B. E. (2008). FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Reviews. Cancer, 8, 83–93.
Yada, M., Hatakeyama, S., Kamura, T., et al. (2004). Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO Journal, 23, 2116–2125.
Strohmaier, H., Spruck, C. H., Kaiser, P., et al. (2001). Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature, 413, 316–322.
Nateri, A. S., Riera-Sans, L., Da Costa, C., & Behrens, A. (2004). The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science, 303, 1374–1378.
Oberg, C., Li, J., Pauley, A., et al. (2001). The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. Journal of Biological Chemistry, 276, 35847–35853.
Inuzuka, H., Shaik, S., Onoyama, I., et al. (2011). SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature, 471, 104–109.
Mao, J. H., Kim, I. J., Wu, D., et al. (2008). FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science, 321, 1499–1502.
Tan, Y., Sangfelt, O., & Spruck, C. (2008). The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Letters, 271, 1–12.
Maser, R. S., Choudhury, B., Campbell, P. J., et al. (2007). Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature, 447, 966–971.
Mao, J. H., Perez-Losada, J., Wu, D., et al. (2004). Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature, 432, 775–779.
Matsuoka, S., Oike, Y., Onoyama, I., et al. (2008). Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes & Development, 22, 986–991.
Moberg, K. H., Bell, D. W., Wahrer, D. C., Haber, D. A., & Hariharan, I. K. (2001). Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature, 413, 311–316.
Kemp, Z., Rowan, A., Chambers, W., et al. (2005). CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Research, 65, 11361–11366.
Calhoun, E. S., Jones, J. B., Ashfaq, R., et al. (2003). BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. American Journal of Pathology, 163, 1255–1260.
Akhoondi, S., Sun, D., von der Lehr, N., et al. (2007). FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Research, 67, 9006–9012.
Skaar, J. R., Pagan, J. K., & Pagano, M. (2009). SnapShot: F box proteins I. Cell, 137(1160–1160), e1161.
Skaar, J. R., D’Angiolella, V., Pagan, J. K., & Pagano, M. (2009). SnapShot: F box proteins II. Cell, 137, 1358. 1358.e1.
Ho, M. S., Tsai, P. I., & Chien, C. T. (2006). F-box proteins: the key to protein degradation. Journal of Biomedical Science, 13, 181–191.
Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J., & Harper, J. W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell, 91, 209–219.
Spruck, C. H., Strohmaier, H., Sangfelt, O., et al. (2002). hCDC4 gene mutations in endometrial cancer. Cancer Research, 62, 4535–4539.
Grim, J. E., Gustafson, M. P., Hirata, R. K., et al. (2008). Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. The Journal of Cell Biology, 181, 913–920.
Matsumoto, A., Tateishi, Y., Onoyama, I., et al. (2011). Fbxw7beta resides in the endoplasmic reticulum membrane and protects cells from oxidative stress. Cancer Science, 102, 749–755.
Welcker, M., Orian, A., Grim, J. E., Eisenman, R. N., & Clurman, B. E. (2004). A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Current Biology, 14, 1852–1857.
Zhang, W., & Koepp, D. M. (2006). Fbw7 isoform interaction contributes to cyclin E proteolysis. Molecular Cancer Research, 4, 935–943.
Bai, C., Sen, P., Hofmann, K., et al. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86, 263–274.
Perkins, G., Drury, L. S., & Diffley, J. F. (2001). Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. The EMBO Journal, 20, 4836–4845.
Orlicky, S., Tang, X., Willems, A., Tyers, M., & Sicheri, F. (2003). Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell, 112, 243–256.
Hao, B., Oehlmann, S., Sowa, M. E., Harper, J. W., & Pavletich, N. P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular Cell, 26, 131–143.
Nash, P., Tang, X., Orlicky, S., et al. (2001). Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature, 414, 514–521.
Welcker, M., & Clurman, B. E. (2007). Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div, 2, 7.
Tang, X., Orlicky, S., Lin, Z., et al. (2007). Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell, 129, 1165–1176.
Pawar, S. A., Sarkar, T. R., Balamurugan, K., et al. (2010). C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proceedings of the National Academy of Sciences of the United States of America, 107, 9210–9215.
Balamurugan, K., Wang, J. M., Tsai, H. H., et al. (2010). The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. The EMBO Journal, 29, 4106–4117.
Strimpakos, A. S., Karapanagiotou, E. M., Saif, M. W., & Syrigos, K. N. (2009). The role of mTOR in the management of solid tumors: an overview. Cancer Treatment Reviews, 35, 148–159.
Isobe, T., Hattori, T., Kitagawa, K., et al. (2009). Adenovirus E1A inhibits SCF(Fbw7) ubiquitin ligase. Journal of Biological Chemistry, 284, 27766–27779.
Koo, E. H., & Kopan, R. (2004). Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nature Medicine, 10(Suppl), S26–S33.
De Strooper, B., Annaert, W., Cupers, P., et al. (1999). A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature, 398, 518–522.
Rocher-Ros, V., Marco, S., Mao, J. H., et al. (2010). Presenilin modulates EGFR signaling and cell transformation by regulating the ubiquitin ligase Fbw7. Oncogene, 29, 2950–2961.
Kim, J., & Bartel, D. P. (2009). Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nature Biotechnology, 27, 472–477.
Xu, Y., Sengupta, T., Kukreja, L., & Minella, A. C. (2010). MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. Journal of Biological Chemistry, 285, 34439–34446.
Lerner, M., Lundgren, J., Akhoondi, S., et al. (2011). MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle, 10, 2172–2183.
Mo, J. S., Ann, E. J., Yoon, J. H., et al. (2011). Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. Journal of Cell Science, 124, 100–112.
BelAiba, R. S., Djordjevic, T., Bonello, S., et al. (2006). The serum- and glucocorticoid-inducible kinase Sgk-1 is involved in pulmonary vascular remodeling: role in redox-sensitive regulation of tissue factor by thrombin. Circulation Research, 98, 828–836.
Kinugawa, K., Yonekura, K., Ribeiro, R. C., et al. (2001). Regulation of thyroid hormone receptor isoforms in physiological and pathological cardiac hypertrophy. Circulation Research, 89, 591–598.
Schulein, C., Eilers, M., & Popov, N. (2011). PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS Letters, 585, 2151–2157.
Durgan, J., & Parker, P. J. (2010). Regulation of the tumour suppressor Fbw7alpha by PKC-dependent phosphorylation and cancer-associated mutations. Biochemistry Journal, 432, 77–87.
Evans, T., Rosenthal, E. T., Youngblom, J., Distel, D., & Hunt, T. (1983). Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell, 33, 389–396.
Harper, J. W., Burton, J. L., & Solomon, M. J. (2002). The anaphase-promoting complex: it’s not just for mitosis any more. Genes & Development, 16, 2179–2206.
Castro, A., Bernis, C., Vigneron, S., Labbe, J. C., & Lorca, T. (2005). The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene, 24, 314–325.
Nakayama, K. I., & Nakayama, K. (2006). Ubiquitin ligases: cell-cycle control and cancer. Nature Reviews. Cancer, 6, 369–381.
Hartwell, L. H., Mortimer, R. K., Culotti, J., & Culotti, M. (1973). Genetic control of the cell division cycle in yeast: V. Genetic Analysis of cdc Mutants. Genetics, 74, 267–286.
Hubbard, E. J., Wu, G., Kitajewski, J., & Greenwald, I. (1997). sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes & Development, 11, 3182–3193.
Koepp, D. M., Schaefer, L. K., Ye, X., et al. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science, 294, 173–177.
Welcker, M., Orian, A., Jin, J., et al. (2004). The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America, 101, 9085–9090.
Wei, W., Jin, J., Schlisio, S., Harper, J. W., & Kaelin, W. G., Jr. (2005). The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell, 8, 25–33.
Ishikawa, Y., Onoyama, I., Nakayama, K. I., & Nakayama, K. (2008). Notch-dependent cell cycle arrest and apoptosis in mouse embryonic fibroblasts lacking Fbxw7. Oncogene, 27, 6164–6174.
Zhao, D., Zheng, H. Q., Zhou, Z., & Chen, C. (2010). The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Research, 70, 4728–4738.
Clurman, B. E., Sheaff, R. J., Thress, K., Groudine, M., & Roberts, J. M. (1996). Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes & Development, 10, 1979–1990.
Won, K. A., & Reed, S. I. (1996). Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. The EMBO Journal, 15, 4182–4193.
Spruck, C. H., Won, K. A., & Reed, S. I. (1999). Deregulated cyclin E induces chromosome instability. Nature, 401, 297–300.
Minella, A. C., Grim, J. E., Welcker, M., & Clurman, B. E. (2007). p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene, 26, 6948–6953.
Ye, X., Nalepa, G., Welcker, M., et al. (2004). Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. Journal of Biological Chemistry, 279, 50110–50119.
Eilers, M., Schirm, S., & Bishop, J. M. (1991). The MYC protein activates transcription of the alpha-prothymosin gene. The EMBO Journal, 10, 133–141.
Bahram, F., von der Lehr, N., Cetinkaya, C., & Larsson, L. G. (2000). c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood, 95, 2104–2110.
Grandori, C., Cowley, S. M., James, L. P., & Eisenman, R. N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annual Review of Cell and Developmental Biology, 16, 653–699.
Adhikary, S., & Eilers, M. (2005). Transcriptional regulation and transformation by Myc proteins. Nature Reviews Molecular Cell Biology, 6, 635–645.
Hann, S. R., & Eisenman, R. N. (1984). Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Molecular and Cellular Biology, 4, 2486–2497.
Amati, B. (2004). Myc degradation: dancing with ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America, 101, 8843–8844.
Salghetti, S. E., Muratani, M., Wijnen, H., Futcher, B., & Tansey, W. P. (2000). Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proceedings of the National Academy of Sciences of the United States of America, 97, 3118–3123.
Flinn, E. M., Busch, C. M., & Wright, A. P. (1998). myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Molecular and Cellular Biology, 18, 5961–5969.
Sears, R., Nuckolls, F., Haura, E., et al. (2000). Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & Development, 14, 2501–2514.
Popov, N., Schulein, C., Jaenicke, L. A., & Eilers, M. (2010). Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nature Cell Biology, 12, 973–981.
Hartl, M., Bader, A. G., & Bister, K. (2003). Molecular targets of the oncogenic transcription factor jun. Current Cancer Drug Targets, 3, 41–55.
Behrens, A., Sibilia, M., & Wagner, E. F. (1999). Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nature Genetics, 21, 326–329.
Shaulian, E., Schreiber, M., Piu, F., et al. (2000). The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell, 103, 897–907.
Szabowski, A., Maas-Szabowski, N., Andrecht, S., et al. (2000). c-Jun and JunB antagonistically control cytokine-regulated mesenchymal–epidermal interaction in skin. Cell, 103, 745–755.
Fuchs, S. Y., Xie, B., Adler, V., et al. (1997). c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. Journal of Biological Chemistry, 272, 32163–32168.
Musti, A. M., Treier, M., & Bohmann, D. (1997). Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science, 275, 400–402.
Radtke, F., Schweisguth, F., & Pear, W. (2005). The Notch ‘gospel’. EMBO Reports, 6, 1120–1125.
Artavanis-Tsakonas, S., Rand, M. D., & Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776.
Demarest, R. M., Ratti, F., & Capobianco, A. J. (2008). It’s T-ALL about Notch. Oncogene, 27, 5082–5091.
Radtke, F., Wilson, A., Mancini, S. J., & MacDonald, H. R. (2004). Notch regulation of lymphocyte development and function. Nature Immunology, 5, 247–253.
Gupta-Rossi, N., Le Bail, O., Gonen, H., et al. (2001). Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. Journal of Biological Chemistry, 276, 34371–34378.
Wu, G., Lyapina, S., Das, I., et al. (2001). SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Molecular and Cellular Biology, 21, 7403–7415.
Wu, G., Hubbard, E. J., Kitajewski, J. K., & Greenwald, I. (1998). Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proceedings of the National Academy of Sciences of the United States of America, 95, 15787–15791.
Fryer, C. J., White, J. B., & Jones, K. A. (2004). Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Molecular Cell, 16, 509–520.
O’Neil, J., Grim, J., Strack, P., et al. (2007). FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. The Journal of Experimental Medicine, 204, 1813–1824.
Thompson, B. J., Buonamici, S., Sulis, M. L., et al. (2007). The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. The Journal of Experimental Medicine, 204, 1825–1835.
Weng, A. P., Ferrando, A. A., Lee, W., et al. (2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science, 306, 269–271.
Girard, L., Hanna, Z., Beaulieu, N., et al. (1996). Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes & Development, 10, 1930–1944.
Feldman, B. J., Hampton, T., & Cleary, M. L. (2000). A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A-PBX1 transgenic mice. Blood, 96, 1906–1913.
Beverly, L. J., & Capobianco, A. J. (2003). Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell, 3, 551–564.
Rohn, J. L., Lauring, A. S., Linenberger, M. L., & Overbaugh, J. (1996). Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. Journal of Virology, 70, 8071–8080.
Yan, X. Q., Sarmiento, U., Sun, Y., et al. (2001). A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood, 98, 3793–3799.
Dorsch, M., Zheng, G., Yowe, D., et al. (2002). Ectopic expression of Delta4 impairs hematopoietic development and leads to lymphoproliferative disease. Blood, 100, 2046–2055.
Kuiperij, H. B., van der Horst, A., Raaijmakers, J., et al. (2005). Activation of FoxO transcription factors contributes to the antiproliferative effect of cAMP. Oncogene, 24, 2087–2095.
Fan, X., Matsui, W., Khaki, L., et al. (2006). Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Research, 66, 7445–7452.
Fernandez-Majada, V., Aguilera, C., Villanueva, A., et al. (2007). Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America, 104, 276–281.
Moriyama, M., Osawa, M., Mak, S. S., et al. (2006). Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. The Journal of Cell Biology, 173, 333–339.
Miyamoto, Y., Maitra, A., Ghosh, B., et al. (2003). Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell, 3, 565–576.
Sriuranpong, V., Borges, M. W., Ravi, R. K., et al. (2001). Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Research, 61, 3200–3205.
Nicolas, M., Wolfer, A., Raj, K., et al. (2003). Notch1 functions as a tumor suppressor in mouse skin. Nature Genetics, 33, 416–421.
Qi, R., An, H., Yu, Y., et al. (2003). Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Research, 63, 8323–8329.
Nguyen, B. C., Lefort, K., Mandinova, A., et al. (2006). Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes & Development, 20, 1028–1042.
Onoyama, I., Tsunematsu, R., Matsumoto, A., et al. (2007). Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. The Journal of Experimental Medicine, 204, 2875–2888.
Liu, Y., Wen, J. K., Dong, L. H., Zheng, B., & Han, M. (2010). Kruppel-like factor (KLF) 5 mediates cyclin D1 expression and cell proliferation via interaction with c-Jun in Ang II-induced VSMCs. Acta Pharmacologica Sinica, 31, 10–18.
Nandan, M. O., Chanchevalap, S., Dalton, W. B., & Yang, V. W. (2005). Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Letters, 579, 4757–4762.
Chen, C., Benjamin, M. S., Sun, X., et al. (2006). KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. International Journal of Cancer, 118, 1346–1355.
Zheng, H. Q., Zhou, Z., Huang, J., et al. (2009). Kruppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene, 28, 3702–3713.
Yagi, N., Manabe, I., Tottori, T., et al. (2009). A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Research, 69, 6531–6538.
Chen, C., Zhou, Z., Guo, P., & Dong, J. T. (2007). Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Letters, 581, 1124–1130.
Oh, I. H., & Reddy, E. P. (1999). The myb gene family in cell growth, differentiation and apoptosis. Oncogene, 18, 3017–3033.
Slamon, D. J., Boone, T. C., Murdock, D. C., et al. (1986). Studies of the human c-myb gene and its product in human acute leukemias. Science, 233, 347–351.
Siegert, W., Beutler, C., Langmach, K., Keitel, C., & Schmidt, C. A. (1990). Differential expression of the oncoproteins c-myc and c-myb in human lymphoproliferative disorders. European Journal of Cancer, 26, 733–737.
Kitagawa, K., Hiramatsu, Y., Uchida, C., et al. (2009). Fbw7 promotes ubiquitin-dependent degradation of c-Myb: involvement of GSK3-mediated phosphorylation of Thr-572 in mouse c-Myb. Oncogene, 28, 2393–2405.
Kanei-Ishii, C., Nomura, T., Takagi, T., et al. (2008). Fbxw7 acts as an E3 ubiquitin ligase that targets c-Myb for nemo-like kinase (NLK)-induced degradation. Journal of Biological Chemistry, 283, 30540–30548.
Kern, S. E., Kinzler, K. W., Bruskin, A., et al. (1991). Identification of p53 as a sequence-specific DNA-binding protein. Science, 252, 1708–1711.
Kimura, T., Gotoh, M., Nakamura, Y., & Arakawa, H. (2003). hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Science, 94, 431–436.
Finkin, S., Aylon, Y., Anzi, S., Oren, M., & Shaulian, E. (2008). Fbw7 regulates the activity of endoreduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene, 27, 4411–4421.
Flores, E. R., Sengupta, S., Miller, J. B., et al. (2005). Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell, 7, 363–373.
Yang, A., Schweitzer, R., Sun, D., et al. (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature, 398, 714–718.
Mills, A. A., Zheng, B., Wang, X. J., et al. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature, 398, 708–713.
Thurfjell, N., Coates, P. J., Uusitalo, T., et al. (2004). Complex p63 mRNA isoform expression patterns in squamous cell carcinoma of the head and neck. International Journal of Oncology, 25, 27–35.
van Bokhoven, H., & McKeon, F. (2002). Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends in Molecular Medicine, 8, 133–139.
Laurikkala, J., Mikkola, M. L., James, M., et al. (2006). p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development, 133, 1553–1563.
Rossi, M., De Simone, M., Pollice, A., et al. (2006). Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle, 5, 1816–1822.
Rossi, M., Aqeilan, R. I., Neale, M., et al. (2006). The E3 ubiquitin ligase Itch controls the protein stability of p63. Proceedings of the National Academy of Sciences of the United States of America, 103, 12753–12758.
Galli, F., Rossi, M., D’Alessandra, Y., et al. (2010). MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. Journal of Cell Science, 123, 2423–2433.
Massague, J., & Wotton, D. (2000). Transcriptional control by the TGF-beta/Smad signaling system. The EMBO Journal, 19, 1745–1754.
Miyazono, K., Suzuki, H., & Imamura, T. (2003). Regulation of TGF-beta signaling and its roles in progression of tumors. Cancer Science, 94, 230–234.
Bengoechea-Alonso, M. T., & Ericsson, J. (2010). Tumor suppressor Fbxw7 regulates TGFbeta signaling by targeting TGIF1 for degradation. Oncogene, 29, 5322–5328.
Besirli, C. G., Wagner, E. F., & Johnson, E. M., Jr. (2005). The limited role of NH2-terminal c-Jun phosphorylation in neuronal apoptosis: identification of the nuclear pore complex as a potential target of the JNK pathway. The Journal of Cell Biology, 170, 401–411.
Hoeck, J. D., Jandke, A., Blake, S. M., et al. (2010). Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nature Neuroscience, 13, 1365–1372.
Matsumoto, A., Onoyama, I., Sunabori, T., et al. (2011). Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. Journal of Biological Chemistry, 286, 13754–13764.
Willis, S. N., Fletcher, J. I., Kaufmann, T., et al. (2007). Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science, 315, 856–859.
Wertz, I. E., Kusam, S., Lam, C., et al. (2011). Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature, 471, 110–114.
Sporn, M. B. (1996). The war on cancer. Lancet, 347, 1377–1381.
Howe, A., Aplin, A. E., Alahari, S. K., & Juliano, R. L. (1998). Integrin signaling and cell growth control. Current Opinion in Cell Biology, 10, 220–231.
Coussens, L. M., & Werb, Z. (1996). Matrix metalloproteinases and the development of cancer. Chemistry & Biology, 3, 895–904.
Chambers, A. F., & Matrisian, L. M. (1997). Changing views of the role of matrix metalloproteinases in metastasis. Journal of the National Cancer Institute, 89, 1260–1270.
Wolfer, A., & Ramaswamy, S. (2011). MYC and metastasis. Cancer Research, 71, 2034–2037.
Massague, J., Blain, S. W., & Lo, R. S. (2000). TGFbeta signaling in growth control, cancer, and heritable disorders. Cell, 103, 295–309.
Knuutila, S., Aalto, Y., Autio, K., et al. (1999). DNA copy number losses in human neoplasms. American Journal of Pathology, 155, 683–694.
Rajagopalan, H., Jallepalli, P. V., Rago, C., et al. (2004). Inactivation of hCDC4 can cause chromosomal instability. Nature, 428, 77–81.
Woo Lee, J., Hwa Soung, Y., Young Kim, S., et al. (2006). Somatic mutation of hCDC4 gene is rare in lung adenocarcinomas. Acta Oncologica, 45, 487–488.
Nowak, D., Mossner, M., Baldus, C. D., et al. (2006). Mutation analysis of hCDC4 in AML cells identifies a new intronic polymorphism. International Journal of Medical Sciences, 3, 148–151.
Kwak, E. L., Moberg, K. H., Wahrer, D. C., et al. (2005). Infrequent mutations of Archipelago (hAGO, hCDC4, Fbw7) in primary ovarian cancer. Gynecologic Oncology, 98, 124–128.
Yan, T., Wunder, J. S., Gokgoz, N., et al. (2006). hCDC4 variation in osteosarcoma. Cancer Genetics and Cytogenetics, 169, 138–142.
Fresno Vara, J. A., Casado, E., de Castro, J., et al. (2004). PI3K/Akt signalling pathway and cancer. Cancer Treatment Reviews, 30, 193–204.
Larson Gedman, A., Chen, Q., Kugel Desmoulin, S., et al. (2009). The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and downstream signaling in pediatric T-cell acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Leukemia, 23, 1417–1425.
Acknowledgments
This work was supported by Canadian Institutes of Health Research (MOP-84559, MOP-93810, and MOP-110974), Canadian Cancer Society Research Institute (2011-700714), and Canadian Dermatology Foundation to G. Li. Y. Cheng is a recipient of the trainee award from Canadian Institute of Health Research Skin Research Training Centre and University of British Columbia Graduate Fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, Y., Li, G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev 31, 75–87 (2012). https://doi.org/10.1007/s10555-011-9330-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-011-9330-z