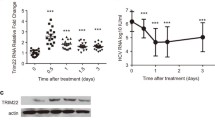
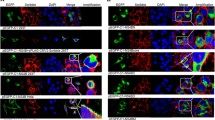
Abstract
The hepatitis C virus (HCV) NS5A gene product is a phosphorylated 56- to 58-kD nonstructural protein that displays a multitude of activities related to enhancement of viral pathogenesis. Although associated with other viral encoded proteins as part of the viral replicase complex positioned on the cytoplasmic side of the endoplasmic reticulum, a role for NS5A in viral replication has not been defined. Post-translational modifications of NS5A include phosphorylation and potential proteolytic processing to smaller molecular weight forms able to translocate to the nucleus. Both the identification of a putative interferon (IFN) sensitivity-determining region within NS5A, as well as the direct interaction with and inhibition of the IFN-induced double-stranded RNA-dependent protein kinase (PKR) by NS5A remain controversial. Truncated versions of NS5A can act as transcriptional activators, while other recently characterized interactions of NS5A with cellular proteins indicate its pleiotropic role in HCV-host interactions. NS5A itself has no direct effect on IFN-α signaling or activation, but other abundant interactions with members of the cellular signaling apparatus, transcription activation machinery and cell cycle-regulatory kinases have been described (e.g. growth factor receptor-bound protein 2, p53, p21/waf and cyclins). Many of these interactions block the apoptotic cellular response to persistent HCV infection. More recently, another altogether different mechanism attenuating the IFN-α response was reported based on induction of interleukin (IL)-8. IL-8, in model systems, potentiates viral replication and mutes the nonspecific intracellular IFN antiviral response. Evidence supporting a complex multimechanistic role of NS5A in promoting viral persistence, pathogenesis and, indirectly, viral-related hepatocarcinogenesis indicates its key role in HCV pathobiology.
Similar content being viewed by others
References
Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: A perspective on long-term outcome. Semin Liver Dis 20:17–35;2000.
Arima N, Kao CY, Licht T, Padmanabhan R, Sasaguri Y, Padmanabhan R. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol Chem 276:12675–12684;2001.
Asabe S-I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of the hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol 71:790–796;1997.
Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J Virol 74:1513–1523;2000.
Blight K, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974;2000.
Choo Q-L, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362;1989.
Chung KM, Song OK, Jang SK. Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol Cells 7:661–667;1997.
David M, Petricoin E, Benjamin C, Pine R, Weber MJ, Larner AC. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science 269:1721–1723;1995.
Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest 96:224–230;1995.
Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med 334:77–81;1996.
Fath I, Schweighoffer F, Rey I, Multon MC, Boiziau J, Duchesne M, Tocque B. Cloning of a Grb2 isoform with apoptotic properties. Science 264:971–974;1994.
François C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum HE, Gatignol A, Wychowski C, Moradpour D, Meurs EF. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol 74:5587–5596;2000.
Fretz H, Furet P, Garcia-Echeverria C, Schoepfer J, Rahuel J. Structure-based design of compounds inhibiting Grb2-SH2 mediated protein-protein interactions in signal transduction pathways. Curr Pharm Des 6:1777–1796;2000.
Fukuma T, Enomoto N, Marumo F, Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology 28:1147–1153;1998.
Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217–227;1997.
Gale M Jr, Kwieciszewski B, Dossett M, Nakao H, Katze MG. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol 73:6506–6516;1999.
Gao T, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MMC. Hepatitis C virus RNA polymerase NS5A complex with a SNARE-like protein. Virology 263:30–41;1999.
Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res 67:173–178;2000.
Ghosh AK, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J Biol Chem 275:7184–7188;2000.
Ghosh AKR, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol 80:1179–1183;1999.
Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc Natl Acad Sci USA 88:9599–9604;2001.
Guo J-T, Bichko VV, Seeger C. Effects of alpha interferon on the hepatitis C virus replicon. J Virol 75:8516–8523;2001.
He Y, Tan SL, Tareen SU, Vijaysri S, Langland JO, Jacobs BL, Katze MG. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J Virol 75:5090–5098;2001.
Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol 73:8469–8475;1999.
Herion D, Hoofnagle JH. The interferon sensitivity determining region: All hepatitis C virus isolates are not the same. Hepatology 25:769–771;1997.
Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA 90:10773–10777;1993.
Hirota M, Satoh S, Asabe S, Kohara M, Tsukiyama-Kohara K, Kato N, Hijikata M, Shimotohno K. Phosphorylation of nonstructural 5A protein of hepatitis C virus: HCV group-specific hyperphosphorylation. Virology 257:130–137;1999.
Hu K-E, Vierling JM, Redeker AG. Viral, host and interferon related factors modulating the effect of interferon therapy for hepatitis C virus infection. J Viral Hepat 8:1–18;2001.
Hummer BT, Li X-L, Hassel BA. Role of p53 in gene induction by double-stranded RNA. J Virol 75:7774–7777;2001.
Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-α catalytic subunit. Gene 201:151–158;1997.
Ide Y, Zhang L, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C nonstructural protein NS5A. Gene 182:203–211;1996.
Jaeckel E, Cornberg M, Wedemeyer H, Santantonio T, Mayer J, Zankel M, Pastore G, Dietrich M, Trautwein C, Manns M. Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med 345:1452–1457;2001.
Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem 274:16370–16376;1999.
Kaneko T, Tanji Y, Satoh S, Hijikata M, Sinichi S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun 205:320–326;1994.
Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem 270:19100–19106;1995.
Kato N. Genome of human hepatitis C virus (HCV): Gene organization, sequence diversity and variation. Microb Comp Genomics 5:129–151;2000.
Kato N, Lan K-H, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol 71:8856–8859;1997.
Katze MG, Kwieciszewski B, Goodlet DR, Blakely CM, Nedderman P, Tan S-L, Aebersold R. Ser2194 is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology 278:501–513;2000.
Khabar KSA, Al-Zoghaibi F, Al-Ahdal MN, Murayama T, Dhalla M, Mukaida N, Taha M, Al-Sedairy ST, Siddiqui Y, Kessie G, Matsushima K. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon α. J Exp Med 186:1077–1085;1997.
Khabar KS, Al-Zoghaibi F, Murayama T, Matsushima K, Mukaida N, Siddiqui Y, Dhalla M, Al-Ahdal MN. Interleukin-8 selectively enhances cytopathic effect (CPE) induced by positive-strand RNA viruses in the human WISH cell line. Biochem Biophys Res Commun 235:774–781;1997.
Kim J, Lee D, Choe J. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem Biophys Res Commun 257:777–781;1999.
Lane BR, Lore K, Bock PJ, Andersson J, Coffey MJ, Strieter RM, Markovitz DM. Interleukin-8 stimulates human immunodeficiency virus type 1 replication and is a potential target for antiretroviral therapy. J Virol 75:5812–5822;2001.
Lemon SM, Lerat H, Weinman SA, Honda M. A transgenic mouse model of steatosis and hepatocellular carcinoma associated with chronic hepatitis C virus infection in humans. Trans Am Clin Climatol Assoc 111:146–157;2000.
Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 75:1437–1449;2001.
Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113;1999.
Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol 75:1401–1407;2001.
Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 358:958–965;2001.
Miller RH, Purcell RH. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA 87:2057–2061;1990.
Mukaida N, Harada A, Matsushima K. Interleukin-8 (IL-8) and monocyte chemotactic and activating factor (MCAF/MCP-1), chemokines essentially involved in inflammatory and immune reactions. Cytokine Growth Factor Rev 9:9–23;1998.
Murayama T, Ohara Y, Obuchi M, Khabar KSA, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1 and NF-κB-binding sites of the interleukin-8 gene. J Virol 71:5692–5695;1997.
Nakamura H, Li M, Zarycki J, Jung JU. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 75:7572–7582;2001.
Neddermann P, Clementi A, De Francesco R. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B and NS5A encoded on the same polyprotein. J Virol 73:9984–9991;1999.
Neuman MG, Benhamou J-P, Malkiewicz IM, Akremi R, Shear NH, Asselah T, Ibrahim A, Boyer N, Martinot-Peignoux M, Jacobson-Brown P, Katz GG, Le Breton V, Le Guludec G, Suneja A, Marcellin P. Cytokines as predictors for sustained response and as markers for immunomodulation in patients with chronic hepatitis C. Clin Biochem 34:173–182;2001.
Parcells MS, Lin S-F, Dienglewicz RL, Majerciak V, Robinson DR, Chen H-C, Wu Z, Dubyak GR, Brunovskis P, Hunt HD, Lee LF, Kung H-J. Marek's disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): Characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J Virol 75:5159–5173;2001.
Pawlotsky JM, Germanidis G. The nonstructural 5A protein of hepatitis C virus. J Viral Hepat 6:343–356;1999.
Podevin P, Sabile A, Gajardo R, Delheim N, Abadie A, Lozach P-Y, Beretta L, Brechot C. Expression of hepatitis C virus NS5A natural mutants in a cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology 33:1503–1511;2001.
Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol 75:6095–6106;2001.
Polyak SJ, Khabar KS, Rezeiq M, Gretch DR. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol 75:6209–6211;2001.
Reed KE, Rice CM. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J Biol Chem 274:28011–28018;1999.
Reed KE, Xu J, Rice CM. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: Properties of the NS5A-associated kinase. J Virol 71:7187–7197;1997.
Satoh S, Hirota M, Noguchi T, Hijikata M, Handa H, Shimotohno K. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology 270:476–487;2000.
Song J, Nagano-Fujii M, Wang F, Florese R, Fujita T, Ishido S, Hotta H. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res 69:109–117;2000.
Sun Y. Free radicals, antioxidant enzymes, and carcinogenesis. Free Radic Biol Med 8:583–599;1990.
Tai D, Tsai S, Chen Y, Chuang Y, Peng C, Sheen I, Yeh C, Chang K, Huang S, Kuo, G Liaw Y. Activation of nuclear factor kappa B in hepatitis C virus infection: Implications for pathogenesis and hepatocarcinogenesis. Hepatology 31:656–664;2000.
Takada S, Kaneniwa N, Tsuchida N, Koike K. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene 15:1895–1901;1997.
Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: The jury is still out on NS5A. Virology 284:1–12;2001.
Tan S-L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs BL, Mayer BJ, Katze MG. NS5A, a nonstructural protein of the hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA 96:5533–5538;1999.
Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun 236:360–364;1997.
Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus encoded nonstructural protein NS5A. J Virol 69:3980–3986;1995.
Taylor DR. Hepatitis C virus and interferon resistance: It's more than just PKR. Hepatology 33:1547–1549;2001.
Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107–110;1999.
Terrault NA. Hepatitis C virus and liver transplantation. Semin Gastrointest Dis 11:96–114;2000.
Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 408:307–310;2000.
Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710–721;2001.
Wasley A, Alter MJ. Epidemiology of hepatitis C: Geographic differences and temporal trends. Semin Liver Dis 20:1–16;2000.
Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J 20:3840–3848;2001.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Reyes, G.R. The nonstructural NS5A protein of hepatitis C virus: An expanding, multifunctional role in enhancing hepatitis C virus pathogenesis. J Biomed Sci 9, 187–197 (2002). https://doi.org/10.1007/BF02256065
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02256065