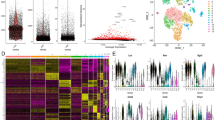
Abstract
Pancreatic stellate cells (PSCs) are thought to be the primary source of the extensive fibrotic reaction characteristic of pancreatic cancer and chronic pancreatitis in humans. PSCs share many morphological and functional characteristics with hepatic stellate cells (HSCs), whose central role in liver fibrosis is well established. However, it has remained unclear if hepatic and pancreatic stellate cells are derived from a common cell lineage and if they are completely similar or if they possess organ-specific features. We have analysed the transcriptomes of HSCs, PSCs and skin fibroblasts to assess how the transcriptional phenotype of stellate cells differs from that of a typical fibroblast lineage cell and if there is evidence for a common stellate cell precursor. To this end, we have performed expression profiling of primary cultures of human HSCs, PSCs and skin fibroblasts using 23,000-feature ‘whole genome’ oligonucleotide micro-arrays. Expression data were verified using real-time PCR. The expression profiles of HSCs and PSCs displayed a great extent of similarity, clearly separating them from the fibroblasts. Predominantly extracellular and cell surface genes, but also signalling molecules, transcription factors and novel neural markers, were concordantly expressed in both stellate cell types. Despite this high degree of similarity, distinct differences in expression patterns were observed between HSCs and PSCs, reflecting organ-specific variations of the common stellate cell-specific phenotype.



Similar content being viewed by others
References
Gress TM, Muller-Pillasch F, Lerch MM, Friess H, Buchler M, Adler G (1995) Expression and in situ localization of genes coding for extracellular matrix proteins and extracellular matrix degrading proteases in pancreatic cancer. Int J Cancer 62:407–413
Apte MV, Haber PS, Applegate TL, Norton ID, McCaughan GW, Korsten MA, Pirola RC, Wilson JS (1998) Periacinar stellate shaped cells in rat pancreas: identification, isolation, and culture. Gut 43:128–133
Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grunert A, Adler G (1998) Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 115:421–432
Blomhoff R, Wake K (1991) Perisinusoidal stellate cells of the liver: important roles in retinol metabolism and fibrosis. FASEB J 5:271–277
Friedman SL (1993) Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med 328:1828–1835
Friedman SL (1990) Cellular sources of collagen and regulation of collagen production in liver. Semin Liver Dis 10:20–29
McGee JO, Patrick RS (1972) The role of perisinusoidal cells in experimental hepatic fibrogenesis. J Pathol 106:vi
Gressner AM, Bachem MG (1995) Molecular mechanisms of liver fibrogenesis—a homage to the role of activated fat-storing cells. Digestion 56:335–346
Geerts A (2001) History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis 21:311–335
Ballardini G, Fallani M, Biagini G, Bianchi FB, Pisi E (1988) Desmin and actin in the identification of Ito cells and in monitoring their evolution to myofibroblasts in experimental liver fibrosis. Virchows Arch B Cell Pathol Incl Mol Pathol 56:45–49
Bachem MG, Meyer D, Schafer W, Riess U, Melchior R, Sell KM, Gressner AM (1993) The response of rat liver perisinusoidal lipocytes to polypeptide growth regulator changes with their transdifferentiation into myofibroblast-like cells in culture. J Hepatol 18:40–52
Bachem MG, Sell KM, Melchior R, Kropf J, Eller T, Gressner AM (1993) Tumor necrosis factor alpha (TNF alpha) and transforming growth factor beta 1 (TGF beta 1) stimulate fibronectin synthesis and the transdifferentiation of fat-storing cells in the rat liver into myofibroblasts. Virchows Arch B Cell Pathol Incl Mol Pathol 63:123–130
Apte MV, Haber PS, Darby SJ, Rodgers SC, McCaughan GW, Korsten MA, Pirola RC, Wilson JS (1999) Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut 44:534–541
Luttenberger T, Schmid-Kotsas A, Menke A, Siech M, Beger H, Adler G, Grunert A, Bachem MG (2000) Platelet-derived growth factors stimulate proliferation and extracellular matrix synthesis of pancreatic stellate cells: implications in pathogenesis of pancreas fibrosis. Lab Invest 80:47–55
Ramadori G (1991) The stellate cell (Ito-cell, fat-storing cell, lipocyte, perisinusoidal cell) of the liver. New insights into pathophysiology of an intriguing cell. Virchows Arch B Cell Pathol Incl Mol Pathol 61:147–158
Gressner AM (1991) Time-related distribution profiles of sulfated glycosaminoglycans in cells, cell surfaces, and media of cultured rat liver fat-storing cells. Proc Soc Exp Biol Med 196:307–315
Pinzani M (2002) PDGF and signal transduction in hepatic stellate cells. Front Biosci 7:d1720–d1726
Pinzani M, Marra F (2001) Cytokine receptors and signaling in hepatic stellate cells. Semin Liver Dis 21:397–416
Pinzani M, Gesualdo L, Sabbah GM, Abboud HE (1989) Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells. J Clin Invest 84:1786–1793
Mews P, Phillips P, Fahmy R, Korsten M, Pirola R, Wilson J, Apte M (2002) Pancreatic stellate cells respond to inflammatory cytokines: potential role in chronic pancreatitis. Gut 50:535–541
Niki T, Pekny M, Hellemans K, Bleser PD, Berg KV, Vaeyens F, Quartier E, Schuit F, Geerts A (1999) Class VI intermediate filament protein nestin is induced during activation of rat hepatic stellate cells. Hepatology 29:520–527
Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grunert A, Adler G (1998) Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 115:421–432
Schneiderhan W, Schmid-Kotsas A, Zhao J, Grunert A, Nussler A, Weidenbach H, Menke A, Schmid RM, Adler G, Bachem MG (2001) Oxidized low-density lipoproteins bind to the scavenger receptor, CD36, of hepatic stellate cells and stimulate extracellular matrix synthesis. Hepatology 34:729–737
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3. http://www.bepress.com/sagmb/vol3/iss1/art3
Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN (2003) GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 4:R28
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868
Ross AC (1993) Cellular metabolism and activation of retinoids: roles of cellular retinoid-binding proteins. FASEB J 7:317–327
Napoli JL (1996) Retinoic acid biosynthesis and metabolism. FASEB J 10:993–1001
Tolwinski NS, Wieschaus E (2004) Rethinking WNT signaling. Trends Genet 20:177–181
Wodarz A, Nusse R (1998) Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14:59–88
Hari L, Brault V, Kleber M, Lee HY, Ille F, Leimeroth R, Paratore C, Suter U, Kemler R, Sommer L (2002) Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol 159:867–880
Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y (2004) The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131:2791–2801
Satoh K, Kasai M, Ishidao T, Tago K, Ohwada S, Hasegawa Y, Senda T, Takada S, Nada S, Nakamura T, Akiyama T (2004) Anteriorization of neural fate by inhibitor of beta-catenin and T cell factor (ICAT), a negative regulator of Wnt signaling. Proc Natl Acad Sci U S A 101:8017–8021
Kuhl M, Geis K, Sheldahl LC, Pukrop T, Moon RT, Wedlich D (2001) Antagonistic regulation of convergent extension movements in Xenopus by Wnt/beta-catenin and Wnt/Ca2+ signaling. Mech Dev 106:61–76
Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A (2003) Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem 278:32227–32235
Yao J, Lai E, Stifani S (2001) The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol Cell Biol 21:1962–1972
Cassiman D, Denef C, Desmet VJ, Roskams T (2001) Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology 33:148–158
Knittel T, Aurisch S, Neubauer K, Eichhorst S, Ramadori G (1996) Cell-type-specific expression of neural cell adhesion molecule (N-CAM) in Ito cells of rat liver. Up-regulation during in vitro activation and in hepatic tissue repair. Am J Pathol 149:449–462
Niki T, Pekny M, Hellemans K, Bleser PD, Berg KV, Vaeyens F, Quartier E, Schuit F, Geerts A (1999) Class VI intermediate filament protein nestin is induced during activation of rat hepatic stellate cells. Hepatology 29:520–527
Lardon J, Rooman I, Bouwens L (2002) Nestin expression in pancreatic stellate cells and angiogenic endothelial cells. Histochem Cell Biol 117:535–540
Tsai RY, McKay RD (2000) Cell contact regulates fate choice by cortical stem cells. J Neurosci 20:3725–3735
Seaberg RM, Smukler SR, Kieffer TJ, Enikolopov G, Asghar Z, Wheeler MB, Korbutt G, van der Kooy D (2004) Clonal identification of multipotent precursors from adult mouse pancreas that generate neural and pancreatic lineages. Nat Biotechnol 22:1115–1124
Shinji T, Ujike K, Ochi K, Kusano N, Kikui T, Matsumura N, Emori Y, Seno T, Koide N (2002) Establishment of a novel collagenase perfusion method to isolate rat pancreatic stellate cells and investigation of their gene expression of TGF-beta1, type I collagen, and CTGF in primary culture or freshly isolated cells. Acta Med Okayama 56:211–218
Acknowledgements
We thank S. Braun, C. Ruhland, K. Lanz and M. de Groot for excellent technical assistance. We are indebted to Dr. Kaufman, Department of Human Genetics, University of Ulm, for providing the short-term cultures of human skin fibroblasts. This work was supported by grants of the DFG to T.M.G. and M.B. (SFB 518, projects B1 and C4) and M.G.B. (SFB 518, project A7) and of the European Union to T.M.G. (QLG1-CT-2002-01196).
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Buchholz and H. A. Kestler made equal contributions to this study and should both be considered first authors
M. G. Bachem and T. M. Gress made equal contributions to this study and should both be considered senior authors
Rights and permissions
About this article
Cite this article
Buchholz, M., Kestler, H.A., Holzmann, K. et al. Transcriptome analysis of human hepatic and pancreatic stellate cells: organ-specific variations of a common transcriptional phenotype. J Mol Med 83, 795–805 (2005). https://doi.org/10.1007/s00109-005-0680-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-005-0680-2