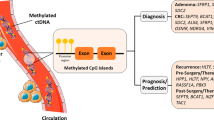
Abstract
Colorectal cancer (CRC) is the third most common type of cancer and is responsible for 9 % of cancer deaths in both men and women in the USA for 2013. It is a heterogenous disease, and its three classification types are microsatellite instability, chromosomal instability, and CpG island methylator phenotype. Biomarkers are molecules, which can be used as indicators of cancer. They have the potential to achieve great sensitivities and specificities in diagnosis and prognosis of CRC. DNA methylation biomarkers are epigenetic markers, more specifically genes that become silenced after aberrant methylation of their promoter in CRC. Some methylation biomarkers like SEPT9 (ColoVantage®) and vimentin (ColoSureTM) are already commercially available. Other blood and fecal-based biomarkers are currently under investigation and clinical studies so that they can be used in the near future. Biomarker panels are also currently being studied since they show great potential in diagnosis as they can combine robust biomarkers to achieve even greater sensitivities than single markers. Finally, methylation-sensitive microRNAs (miRNAs) are very promising markers, and their investigation as biomarkers, is only at primitive stage.


Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30
Brenner H, Bouvier AM, Foschi R, Hackl M, Larsen IK, Lemmens V, Mangone L, Francisci S, The EUROCARE Working Group (2012) Progress in colorectal cancer survival in Europe from the late 1980s to the early 21st century: the EUROCARE study. Int J Cancer 131:1649–1658
Ludwig JA, Weinstein JN (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 5:845–856
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96:8681–8686
Worthley DL, Leggett BA (2010) Colorectal cancer: molecular features and clinical opportunities. Clin Biochem Rev 31:31–38
Derks S, van Engeland M (2012) Promoter CpG island methylation in colorectal cancer. In: Appasani K (ed) Epigenomics: from chromatin biology to therapeutics, 1st edn. Cambridge University Press, Cambridge, pp 307–308
Philips T (2008) The role of methylation in gene expression. Nat Edu 1:127
Laird PW, Jaenisch R (1994) DNA methylation and cancer. Hum Mol Genet 3:1487–1495
Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T (2008) Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 68:4123–4132
Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J (2009) Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 125:2737–2743
Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X (2010) Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 29:5724–5728
Curtin K, Slattery ML, Samowitz WS (2011) CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int 2011:902674. doi:10.4061/2011/902674
Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM et al (2007) The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology 132:127–138
Snover DC (2011) Update on the serrated pathway to colorectal carcinoma. Hum Pathol 42:1–10
Ahn JB, Chung WB, Maeda O, Shin SJ, Kim HS, Chung HC, Kim NK, Issa JP (2011) DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer 117:1847–1854
Kim KM, Lee EJ, Ha S, Kang SY, Jang KT, Park CK, Kim JY, Kim YH, Chang DK, Odze RD (2011) Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol 35:1274–1286
Suzuki H, Igarashi S, Nojima M, Maruyama R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y et al (2010) IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 31:342–349
Dimberg J, Hong TT, Skarstedt M, Löfgren S, Zar N, Matussek A (2013) Analysis of APC and IGFBP7 promoter gene methylation in Swedish and Vietnamese colorectal cancer patients. Oncol Lett 5:25–30
Leggett B, Whitehall V (2010) Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 138:2088–2100
Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS (2007) CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch 450:529–537
Ebert MPA, Tänzer M, Balluff B, Burgermeister E, Kretzschmar AK, Hughes DJ, Tetzner R, Lofton-Day C, Rosenberg R, Reinacher-Schick AC et al (2012) TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med 366:44–53
Lee JH, Kang MJ, Han HY, Lee MG, Jeong SI, Ryu BK, Ha TK, Her NG, Han J, Park SJ et al (2011) Epigenetic alteration of PRKCDBP in colorectal cancers and its implication in tumor cell resistance to TNF-α-induced apoptosis. Clin Cancer Res 17:7551–7562
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138:2073–2087
Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklöf V, Rutegård J, Oberg A, Van Guelpen BR (2010) The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 16:1845–1855
Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23:609–618
Hrašovec S, Glavač D (2012) MicroRNAs as novel biomarkers in colorectal cancer. Front Genet 3:180. doi:10.3389/fgene.2012.00180
Derks S, Postma C, Carvalho B, van den Bosch SM, Moerkerk PT, Herman JG, Weijenberg MP, de Bruïne AP, Meijer GA, van Engeland M (2008) Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis 29:434–439
Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, Fang JC, Samowitz WS, Heichman KA (2011) Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 9:133–141
Tóth K, Sipos F, Kalmar A, Patai AV, Wichmann B, Stoehr R, Golcher H, Scheller V, Tulassay Z, Molnár B (2012) Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 7(11):e48547. doi:10.1371/journal.pone.0046000
Ned RM, Melillo S, Marrone M (2011) Fecal DNA testing for colorectal cancer screening: the ColoSureTM test. PLoS Curr 3:RRN1220. doi:10.1371/currents.RRN1220
Oh T, Kim N, Moon Y, Soon Kim M, Hoehn BD, Hee Park C, Soo Kim T, Kyu Kim N, Cheol Chung H, An S (2013) Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn 15:498–507
Nishio M, Sakakura C, Nagata T, Komiyama S, Miyashita A, Hamada T, Kuryu Y, Ikoma H, Kubota T, Kimura A et al (2010) RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res 30:2673–2682
Wu PP, Zou JH, Tang RN, Yao Y, You CZ (2011) Detection and clinical significance of DLC1 gene methylation in serum DNA from colorectal cancer patients. Chin J Cancer Res 23:283–287
Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT (2011) Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol 106:1110–1118
Melotte V, Lentjes MHFM, van den Bosch SM, Hellenbrekers DMEI, de Hoon JPJ, Wouters KAD, Daenen KLJ, Partouns-Hendriks IEJM, Stressels F, Louwagie J et al (2009) N-myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst 101:916–927
Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruïne AP, Smits KM et al (2009) Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res 69:4691–4699
Hibi K, Goto T, Kitamura YH, Yokomizo K, Sakuraba K, Shirahata A, Mizukami H, Saito M, Ishibashi K, Kigawa G et al (2010) Methylation of TFPI2 gene is frequently detected in advanced well-differentiated colorectal cancer. Anticancer Res 30:1205–1207
Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, Sanada Y (2011) Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett 311:96–100
Takada H, Wakabayashi N, Dohi O, Yasui K, Sakakura C, Mitsufuji S, Taniwaki M, Yoshikawa T (2010) Tissue factor pathway inhibitor 2 (TFPI2) is frequently silenced by aberrant promoter hypermethylation in gastric cancer. Cancer Genet Cytogenet 197:16–24
Hellebrekers DMEI, Lentjes MHFM, van den Bosch SM, Melotte V, Wouters KAD, Daenen KLJ, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S et al (2009) GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res 15:3990–3997
Wang DR, Tang D (2008) Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol 14:524–531
Lind GE, Danielsen SA, Ahlquist T, Merok MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff G et al (2011) Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer 10:85–99
Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP (2012) The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 10:272–277
Carmona FJ, Azuara D, Berenguer-Llergo A, Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R, Villanueva A, Fraga MF et al (2013) DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res 6:656–665
Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH (2009) Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res 15:6185–6191
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127:118–126
Schee K, Fodstad Ø, Flatmark K (2010) MicroRNAs as biomarkers in colorectal cancer. Am J Pathol 177:1592–1599
Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Wilson JKV, Kaz AM et al (2008) Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene 27:3880–3888
Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y et al (2013) SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol 229:12–24
Grosse-Gehling P, Fargeas CA, Dittfeld C, Garbe Y, Alison MR, Corbeil D, Kunz-Schughart LA (2013) CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol 229:355–378
Sakthianandeswaren A, Christie M, D’Andreti C, Tsui C, Jorissen RN, Li S, Fleming NI, Gibbs P, Lipton L, Malaterre J et al (2011) PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res 71:3709–3719
Li X, Yao X, Wang Y, Hu F, Wang F, Jiang L, Liu Y, Wang D, Sun G, Zhao Y (2013) MLH1 promoter methylation frequency in colorectal cancer patients and related clinicopathological and molecular features. PLoS One 8(3):e59064. doi:10.1371/journal.pone.0059064
Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS (2007) Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn 9:305–314
Rawson JB, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, Parfrey PS, Young JP, Pollett A, Green RC et al (2011) Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. Br J Cancer 104:1906–1912
Goto T, Mizukami H, Shirahata A, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, Hibi K (2009) Aberrant methylation of the p16 gene is frequently detected in advanced colorectal cancer. Anticancer Res 29:275–277
Peng H, Long F, Wu Z, Chu Y, Li J, Kuai R, Zhang J, Kang Z, Zhang X, Guan M (2013) Downregulation of DLC-1 gene by promoter methylation during primary colorectal cancer progression. Biomed Res Int 2013:181384. doi:10.1155/2013/181384
Conflict of interest
All authors state that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gyparaki, MT., Basdra, E.K. & Papavassiliou, A.G. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Med 91, 1249–1256 (2013). https://doi.org/10.1007/s00109-013-1088-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1088-z