Abstract
Cancer cell metastasis is a multi-stage process involving invasion into surrounding tissue, intravasation, transit in the blood or lymph, extravasation, and growth at a new site. Many of these steps require cell motility, which is driven by cycles of actin polymerization, cell adhesion and acto-myosin contraction. These processes have been studied in cancer cells in vitro for many years, often with seemingly contradictory results. The challenge now is to understand how the multitude of in vitro observations relates to the movement of cancer cells in living tumour tissue. In this review we will concentrate on actin protrusion and acto-myosin contraction. We will begin by presenting some general principles summarizing the widely-accepted mechanisms for the co-ordinated regulation of actin polymerization and contraction. We will then discuss more recent studies that investigate how experimental manipulation of actin dynamics affects cancer cell invasion in complex environments and in vivo.
Similar content being viewed by others
Actin polymerization drives cancer cell motility
The motility of eukaryotic cells is driven by the polymerization of actin monomers into polarized filaments [1, 2]. These filaments, termed F-actin, are in a constant state of flux with new monomers being added at the ‘barbed’ or ‘plus’ end, and depolymerization at the ‘pointed’ or ‘minus’ end. Actin polymerization can be stimulated in many ways, including increasing the rate of monomer addition to barbed ends, nucleating new filaments, increasing the number of barbed ends, and reducing depolymerization [3]. Our understanding of the molecules involved in regulating these processes has increased dramatically and is summarized in Fig. 1. Stated simply, FH proteins [4] and members of the Ena/VASP family [5] increase the rate of monomer addition to barbed ends. Arp2/3 are components of a multimeric complex that nucleates the formation of new actin filaments, typically from the side of existing filaments [6]. Cofilin can increase the number of barbed ends available for polymerization by severing existing filaments [7]. In motile cells the predominant site of actin polymerization is proximal to the plasma membrane, which is driven forward by the addition of actin monomers. Exactly how actin polymerisation alters the shape of the plasma membrane is unclear; membrane may flow to the front of the cell as result of pushing by polymerising actin filaments or hydrostatic pressure or it may be delivered in vesicles. Although the actin polymerization machinery is not attached to the plasma membrane, many of the regulatory factors are either membrane-anchored small G proteins of the Rho family [8] or phospholipids [9] (Fig. 2). This helps ensure that newly polymerized actin filaments are oriented in the direction of cell migration with their barbed ends directed towards the plasma membrane. The rate of polymerization at barbed ends is also modulated by capping proteins [10], which stearically hinder monomer addition, and by the availability of monomers that are usually maintained in complexes with profilin or thymosin, which are permissive for polymerization but prevent inappropriate polymerization [11].
Patterns of actin polymerization. Actin filaments are polarised with polymerization being catalyzed at the ‘barbed’ or ‘plus’ end (marked with ‘+’). (a) Formin homology (FH) proteins (dimerized green semi-circles) promote actin monomer addition to the barbed end and then move processively with the barbed end as the actin filament is extended. Association of actin filament takes place in FH2 domains. (b) The Arp2/3 complex (pink hexagon) nucleates a new actin filament from the side of an existing one, resulting in an actin branch being formed at a 70° angle to the pre-existing filament. The Arp2/3 complex remains at the branching point, between the side of the pre-existing filament and the pointed end of the new filament. This process may be repeated on the same filament or on newly synthesized filaments. (c) Severing of actin filaments by active cofilin family proteins (red star) results in increased free barbed ends available for actin polymerization, thereby increasing the local density of actin filaments
WASP, WAVE and FH protein complexes. (a) N-WASP in a closed conformation is inactive. A transition to an open conformation may be initiated by binding Cdc42 to the CRIB (Cdc42 and Rac interacting/binding) domain and PIP2 to the basic (B) region. Actin monomers may bind directly to the WH2 (WASP homology 2) domains or via binding to Profilin (Pro) which associates with the proline-rich domain (P-Rich). Arp2/3 associates with the central (C) and acidic (A) domains. The WASP interacting protein (WIP) binds to the WH1 (Wasp homology 1) region where it contributes to the regulation of WASP activity. WAVE also exists in an inactive conformation (not shown). Activation occurs following binding of HSPC300 and a multimeric Abi/Nap/PIR121 complex that is responsive to active Rac. Alternatively, Rac may associate via IRSp53 with the P-Rich region to promote activation. Adaptor proteins such as NCK may associate with NAP125 and/or the P-Rich region to promote activity. Actin binds to a WH2 domain while the Arp2/3 complex associates with the C and A domains. PIP3 binding to the B domain has also been implicated in activation. WHD = Wave homology domain. (b) FH proteins exist in a closed conformation with the carboxyl terminal regions folded back upon the CRIB and FH3 (Formin homology 3) domains. Upon association with GTPases including; RhoA, RhoB, Cdc42 or Rif, FH proteins change conformation to promote actin nucleation and polymerization. Profilin binds to the Formin homology 1 (FH1) domain to provide a source of monomeric actin. Diaphanous interacting proteins (DIP) bind to Formin homology 2 (FH2) domains and stabilize the open active conformation. Filamentous actin also binds to the FH2 domain, which modulates elongation and blocks capping proteins from binding
The activity of the polymerization machinery is very tightly regulated. The Arp2/3 complex is regulated by its association with the WAVE and WASP family of WH2 domain containing proteins (WAVE1, 2, & 3, WASP and N-WASP) that can bind both the Arp2/3 complex and actin monomers (Fig. 2) [6, 12]. This helps to bring actin monomers very close in proximity to the Arp2/3 complex and thereby increases the rate of Arp2/3-mediated actin polymerization. WASP family proteins also bind profilin through poly-proline motifs and this further aids recruitment of actin monomers to the Arp2/3 complex [13]. WH2 domain proteins are themselves subject to very tight regulation through a conformational switch [14, 15]. The VCA domains including the WH2 domain can be masked by intramolecular interactions (Fig. 2); this autoinhibited conformation can be relieved through a range of protein–ligand interactions. Interaction with the GTP-bound form of Cdc42, PIP2 or adaptor molecules such as WIP and Nck, have all been shown to promote the active ‘open’ conformation of WASP or N-WASP [12]. Similarly, interactions with a multimeric complex containing Abi/Nap/PIR121 or IRSp53 can enhance the activity of WAVE proteins, in both cases GTP-bound Rac1 is a key determinant of localization of these complexes [12, 16]. The actin binding protein cortactin also binds to Arp3 and this helps to locate active Arp2/3 complexes to the sides of existing actin filaments leading to branched arrays of F-actin [17].
Like WAVE and WASP family proteins, FH proteins also switch between an auto-inhibited ‘closed’ conformation and an active ‘open’ conformation [6]. Interaction with numerous GTP-binding proteins, including Cdc42, RhoA, RhoB and Rif, and adaptor proteins such as DIP/WISH can stabilise the open conformation thereby promoting actin polymerization driven by the FH2 domain [18–22]. Structural studies indicate that FH2 domains function as dimers with one FH2 domain binding a monomer in the existing actin filament and the other FH2 domain recruiting a new G-actin monomer for polymerisation [23].
These complex regulatory mechanisms serve to ensure that actin is not polymerized inappropriately and that actin polymerization can be increased in response to appropriate stimuli. For example, extracellular cues, including growth factors and extracellular matrix components, control the GTP-loading of Rac1 and Cdc42, the generation and hydrolysis of phospholipids, and the recruitment of adaptor protein complexes to membranes. In addition many regulators of actin polymerisation are phosphorylated in response to external stimuli; in some cases this can have very profound affects on their function (the example of cofilin is discussed below), while in other cases phosphorylation can affect the magnitude of response to other regulatory inputs (for example src-mediated phosphorylation of cortactin) [24]. In addition, although significant attention is paid to the role of Rho and ROCK signalling in the regulation of actin–myosin contraction (see below), this signalling pathway also affects the activity of proteins regulating actin polymerization including cofilin via LIM kinase [25], profilin [26] and FH proteins [27]. These mechanisms enable cells to change shape and move in response to suitable extracellular stimuli during development and in pathological situations such as inflammation and wound healing. However, many of these regulatory pathways also become deregulated in cancer cells (see Table 1) and can contribute to the invasive behaviour of cancers. It should be noted that alterations in pathways that regulate actin dynamics may also affect growth control and cell survival. Therefore, proteins regulating actin dynamics may have been selected for altered expression not only based on their pro-migratory actions, but for a more general cancer-promoting function.
The precise mechanism by which actin polymerization is catalyzed can have marked consequences on the overall F-actin structure produced. Typically, molecules that promote the addition of monomers to barbed ends (FH proteins and Ena/VASP) generate linear F-actin arrays [28]: these are called filopodia if they extend laterally from the cell, or microvilli if they extend dorsally (example of filopodia in Fig. 3). Whereas Arp2/3 mediated actin polymerization commonly results in ‘arc-like’ sheets of F-actin called lamellipodia that extend over the substrate (example of lamellipodia in Fig. 3) (reviewed in [29]). Numerous additional proteins interact with polymerized actin filaments and modulate the geometry and function of the actin structures. Fascin can bundle actin filaments to promote the formation of filopodia and may help to shape branched F-actin networks generated by Arp2/3 nucleation into the parallel arrays of filaments in filopodia [30, 31] (Table 1). Other proteins cross-link actin filaments to form a meshwork or connect the actin network to cell-matrix adhesions and to the plasma membrane. The myosin family of motor proteins can ‘walk’ along actin filaments, either carrying cargoes or, in the case of dimeric myosins, generating contractile force by moving two actin filaments relative to one another [32]. Tropomyosins are actin-binding proteins that recruit myosin to actin filaments and respond to increased calcium concentrations by changing conformation to allow acto-myosin contraction [33].
Diversity of actin organisation in migrating cancer cells. Three different cancer cell lines are shown moving on a 2D substrate: MTLn3 mammary carcinoma cell is shown in the left-hand panel with a broad zone of F-actin at the front, called a lamellipodium, and thick actin cables in the cell body, called stress fibres. BE colon carcinoma cell is shown in the middle panel with prominent ‘ruffled’ zone of F-actin at the front, called a pseudopod, and an elongated morphology. A431 squamous cell carcinoma cell is shown in the right-hand panel with numerous F-actin rich protrusions at the front, called filopodia
Co-operation and plasticity in actin polymerization mechanisms
The molecular machinery that regulates the different facets of actin polymerization functions coordinately in most cell types. For example, cofilin and Arp2/3 co-operate to drive maximal actin polymerization in breast cancer cells. A localized increase in cofilin activity leads to increased numbers of barbed ends for actin polymerization while Arp2/3 promote the nucleation of new filaments [34, 35]. Cofilin activity is localized to a region close the plasma membrane because this is the site where PIP2 is hydrolysed and because of its intrinsic preference for recently polymerised ATP-actin filaments [36], while activation of membrane tethered small G proteins leads to increased Arp2/3 activity at the plasma membrane [37, 38]. It should also be noted that in other contexts (such as if ADP-actin filaments are severed or other co-operating mechanism are not active) cofilin-mediated filament severing can reduce F-actin levels. Numerous separate studies have shown that key actin regulators become deregulated during cancer progression (Table 1), i.e. they appear to be coordinately up-regulated in a sub-set of motile cancer cells [39]. This coordination makes sense if one considers the multi-step and cyclical nature of cell motility; up-regulation of any one of the key regulatory steps in isolation would simply result in other regulators becoming rate-limiting thereby producing little or no overall increase in cell motility. Conversely, disruption of any one regulatory pathway would likely have an effect on motility; there is wealth of literature documenting the effects of disrupting Arp2/3, cofilin, FH proteins and Ena/VASP on cell migration [4–7]. However, if one examines the data in more detail it becomes clear that disruption of any particular actin regulatory mechanism fails to completely abrogate motility, most likely due to compensatory mechanisms maintaining actin polymerization and turn-over, thereby supporting motility, albeit at reduced rates. A particularly striking example, if the generation of branched actin filaments by the Arp2/3 complex is blocked, then extensive filopodia formation is observed which sustains cell motility [40]. Conversely, if the function of Ena/VASP proteins that normally promote filopodia formation is blocked, then cells extend a more persistent and uniform lamellipod leading to increased cell speed [41]. These studies reveal some important principles: that the different mechanisms of actin polymerization co-operate to generate the F-actin structures used for cell motility, and that there is plasticity in the regulation of these mechanisms that enables cells to adapt to interference with any one mechanism. Different relative activities of various regulators of actin polymerisation most likely explain the diverse range of morphologies that can be observed in motile cancer cells (Fig. 3).
Acto-myosin contraction
The ability of cancer cells to move requires force generation to overcome factors that oppose movement (e.g. cell–cell and cell–matrix adhesions, drag, etc.). F-actin assembles with myosin II filaments composed of heavy and regulatory light chains to form a protein complex that uses energy from ATP hydrolysis to power actin–myosin contraction [32, 42]. The resultant generation of contractile force drives the morphological reorganization and extracellular matrix remodelling that facilitate cell movement. Given the profound effects that actin–myosin contractility can have, it is not surprising that there is a sophisticated network of regulatory components that hold a tight rein over this process.
Phosphorylation of the myosin II light chains (MLC) is a key mechanism for regulation of actin–myosin contractility [43]. MLC phosphorylation promotes the release of the myosin heavy chain tail allowing for assembly into filaments, and facilitates the association of the myosin head with F-actin. The myosin head uses ATP to ‘walk’ towards the barbed end. When multimeric myosin is associated with more than one actin filament this causes the filaments to move relative to each another, thereby generating contractile force. MLC phosphorylation has been reported to be mediated by numerous kinases including: the Rho-regulated ROCK1 and ROCK2 [44], the ROCK-regulated ZIPK [45], MRCKα and MRCKβ [46, 47], ILK [48], DAPK 1 [49] and 2 [50], DRAK 1 and 2 [51], PAK [52, 53] and MLCK [54] (Table 1). The ability of these various kinases to phosphorylate MLC allows for multiple signalling pathways to converge on the regulation of actin–myosin contractility. Although it would be difficult to define every condition and cell type in which a specific kinase phosphorylates MLC, studies with small molecule inhibitors indicate that ROCK1 and ROCK2 are the major calcium-independent kinases while MLCK is the major calcium-dependent kinase.
Dephosphorylation of MLC is catalyzed by the PP1M phosphatase complex, which is comprised of a PP1Cδ catalytic subunit, a myosin light chain binding subunit (MBS) and a smaller M20 subunit of unknown function [55]. The MBS is a critical component of the complex as it brings together the phosphatase catalytic subunit with its cognate substrate and because of the role it plays in regulating phosphatase activity. An interesting recent development is the discovery that there are five proteins that may act as the MBS (MYPT1, MYPT2, MYPT3, MBS85 and TIMAP) [56]. The best characterized MBS is the ubiquitously-expressed MYPT1 protein, it appears that the more tissue-restricted MYPT2 likely functions and is regulated similarly [56]. The other MBS proteins have not been studied extensively and their roles in regulating MLC phosphorylation remains to be determined. The major site of MYPT1 phosphorylation is Threonine 696 (numbering relates to the human form), which inhibits phosphatase function [57], possibly by blocking the active site or by disrupting interaction of the catalytic subunit with phosphorylated substrate [58]. Kinases that have been reported to phosphorylate Thr696 include: ROCK1 and ROCK2 [57], MRCKα and MRCKβ [47, 59], ILK [60, 61], ZIPK [62] and the DMPK [63]. Phosphorylation of Threonine 853 by ROCK has also been reported to inhibit MLC dephosphorylation by decreasing MLC binding [57, 64].
MLC phosphorylation is also regulated by the CPI-17 protein [65] (Table 1), which when phosphorylated on Threonine 38 potently inhibits PP1M activity by masking the active site in the catalytic PP1Cδ subunit [66]. A number of the same kinases that phosphorylate MYPT1 have also been shown to phosphorylate CPI-17, including ROCK1 and ROCK2 [67], ZIPK [68] and ILK [69], raising the possibility that kinases which inhibit PP1M activity do so by targeting multiple regulatory proteins. The closely related proteins KEPI and PHI-1 [70, 71] also appear to inhibit PP1C activity in a phosphorylation-dependent manner, but their possible roles in regulating MLC phosphorylation have not been characterized in detail. Elevated expression of CPI-17 in several tumour cell lines has been reported, where inhibition of PP1M led to inactivation of the Merlin tumour suppressor protein and consequent oncogenic transformation [72]. An additional possibility is that elevated CPI-17 expression and/or phosphorylation would contribute to the metastatic ability of tumour cells.
A number of kinases, including ROCK, apparently have two modes for elevating MLC phosphorylation, by acting as direct MLC kinases and by inhibiting PP1M activity. There has not been a great deal of effort spent in trying to dissect the relative contribution of these two pathways to MLC phosphorylation induced by a given kinase. However, one possibility is that the major pathway for some kinases is the phosphorylation of MYPT1 and consequent inhibition of PP1M. As a result, a net gain in MLC phosphorylation would actually require less kinase activity directed towards MLC than under conditions in which PP1M was not inhibited. A manifestation of this effect is the increased calcium sensitivity of MLC phosphorylation and the consequent actin–myosin contractile response that can be induced by ROCK [73]. In this example, it would imply that Ca2+ and/or calcium-regulated kinases such as MLCK or DAPK would cooperate with ROCK to promote contractile force generation, and contribute to metastatic behaviour.
As well as a role in facilitating MLC phosphorylation, calcium may contribute to cancer cell metastasis by binding to proteins such as S100A4 [74]. There is very strong evidence from clinical and experimental studies which indicates a significant role for S100A4 overexpression in increased metastasis and poor prognosis for a wide variety of cancers including; breast, colorectal, pancreatic and renal (Table 1). Intriguingly, S100A4 has an extracellular role in promoting metastasis, possibly by inducing remodelling of the extracellular matrix and/or through interactions with a cell surface receptor, as well as an intracellular role. It has been proposed that S100A4 acts by binding to the myosin II heavy chain [75] and promotes increased directional motility by shifting the balance towards forward protrusions and away from side protrusions [76]. In addition, S100A4 may also affect actin–myosin contractility by direct binding to F-actin [77] and to the actin-binding protein tropomyosin [78].
Tropomyosins are derived from four distinct genes (α, β, γ, δ) that are transcribed and spliced into over 40 isoforms [33, 79]. Although they play key roles in the calcium-responsive contraction of striated muscle, their roles in non-muscle cells are less well defined. Different isoforms appear to have distinct biological functions, as a result the patterns of expression affect how tropomyosins might affect the actin cytoskeleton. The expression of tropomyosin isoforms is frequently altered in tumours (Table 1). Some isoforms appear to recruit myosin to actin filaments [80], and influence the activity of the myosin head ATPase and contractility [81]. Tropomyosin has also been reported to increase actin filament stiffness [82] and protect F-actin from the actions of cofilin [83] and gelsolin [84]. However, some isoforms actually reduce active myosin levels and promote the association of cofilin with actin filaments, resulting in the formation of lamellipodia [80]. To add further complexity, isoforms are sorted to different cellular compartments, and these distributions may change during development or in tumour cells. As a result, actin–myosin regulation may be affected by factors in addition to tropomyosin expression levels. Further research is necessary to determine how both isoform expression and subcellular distribution patterns contribute to tumour cell metastasis.
Coordinating polymerization and contraction
For efficient cell motility the processes of actin polymerization and contraction must be coordinated, with polymerization occurring prior to contraction and more proximal to the plasma membrane. The spatial and temporal controls are linked; the main site of actin polymerization is next to the plasma membrane and as the membrane moves forward the most recently polymerized actin will be adjacent to the membrane with older F-actin further away. Immunofluorescence microscopy of the leading edge of most migrating cells reveals that newly polymerised actin close to the plasma membrane is not associated with the contractile machinery (Fig. 4) [85]. How this spatial and temporal separation is achieved is not clear, here we will propose some mechanisms. The simplest one is that the contractile machinery is not incorporated directly into newly polymerized actin filaments, but binds after being recruited. This would obviously take some time for the components to be assembled depending on the affinities and concentrations of the molecules. For example, F-actin at the extreme leading edge of motile cells is free of tropomyosin, which only becomes associated a few microns back from the plasma membrane [86]. Alternatively, the mechanisms that regulate polymerization and contraction could be coordinated. Small GTPases of the Rho family regulate both the actin polymerization machinery and the contractile machinery. One possibility is that the polymerization and contraction machinery could be activated by the same GTPase, but with different kinetics leading to polymerization preceding contraction. For example, RhoA binds to DRF1 and directly relieves its auto-inhibited conformation to promote actin polymerization [4], whereas the mechanisms by which it promotes acto-myosin contraction and reduces depolymerization or severing of filaments requires ROCK-mediated phosphorylation of various intermediate proteins (e.g. MYPT1 to increase acto-myosin contraction [55] or LIMK to reduce cofilin activity [25]). The consequence of this could be that RhoA activates polymerization very focally and very rapidly, but that contraction is activated more diffusely and slowly. Another possibility is that the polymerization machinery can inhibit some of the regulators of contraction. A major activity of Rac1 in the cell is to promote WAVE dependent actin nucleation [87] (Fig. 2), but it also indirectly inhibits RhoA through the production of ROS [88] which may reduce Rho and ROCK driven contractility during phases of Rac1-driven actin polymerization.
Distinct organisation of F-actin and MLC organisation in a migrating cancer cell. F-actin and pS19-MLC staining of MTLn3 cell are shown in red and green, respectively. Lower right panel shows a line-scan of the intensity of F-actin and pS19-MLC staining in red and green, respectively. Note how the F-actin at the front of the cell is not associated with ‘active’ MLC whereas the F-actin at the rear is. This organisation allows the actin at the front of the cell to extend away from the cell body while the of the cell is pulled towards the middle by thick acto-myosin cables (yellow in merged image)
A common theme in these examples is that precise localized regulation of polymerization and contraction is critical. Excessive or global activation can be as detrimental to motility as lack of activity and this should be considered when attempting to reconcile apparently contradictory findings. For example, both excess and reduced levels of LIMK-mediated phosphorylation of cofilin have been reported to reduce cell motility [25], this could be explained if low levels are required at sites of actin polymerization to allow cofilin to generate new barbed ends for polymerization but high levels are in contractile zones to prevent the severing of filaments required for myosin-mediated contractility [36]. Global LIMK activation would reduce polymerization, while global inactivation would reduce the number of filaments available for the contractile machinery.
Actin organisation in complex environments
Most studies analyzing the generation of filopodia and lamellipodia have used cells cultured on rigid 2D substrates; however, these conditions are clearly different from the environment through which cells move in vivo. Recently, significant effort has been focused on trying to understand how F-actin is organized in cancer cells moving in more complex environments [89]. On thicker substrates composed of matrix proteins, many cancer cells form ventral actin-rich structures called invadopodia that are associated with ECM proteolytic activity [90, 91]. These structures have many similarities with podosomes that are found in cells of monocytic origin. The formation of invadopodia requires the activity of the actin nucleating Arp2/3 complex, regulated by N-WASP and cortactin, and the actin severing action of cofilin [92]. The ability of cancer cells to make invadopodia often correlates with their ability to enter the vasculature [93]. However, the holes generated typically in the ECM by invadopodia (1–2 microns) are small compared to the size of the cell, and cancer cells have not been observed to move through the areas of matrix degradation produced by invadopodia. This may merely reflect a limitation of the experimental systems used, but until this issue is resolved the relationship between invadopodia and cancer cell invasion through matrix barriers will remain a topic of lively debate.
The behaviour of cancer cells cultured in a truly 3D matrix is different from when they are cultured on 2D substrates: F-actin structures like lamellopodia are rarely observed without a planar substrate, and the distinction between the ventral and dorsal surfaces is lost [94] (note the contrasting morphologies in Fig. 5). Cancer cells can be observed moving in 3D matrices with morphologies ranging from very elongated to rounded [89]. The matrix composition and density can also modulate cell motility and we are still learning how best to model tissues architectures in vitro (also discussed in ‘Actin dynamics in living tumours’ section). In most cases there is a zone of actin polymerization of variable size at the front of the cell, which is often rather loosely termed a pseudopod (Fig. 5). The exact relationship between a pseudopod in a 3D matrix and an invadopodium is not entirely clear, although by definition an invadopodium is associated with proteolytic activity [91]. Resolution of this relationship would require simultaneous analysis of actin polymerization and ECM proteolysis. A recent study did examine these processes found that proteolytic activity is restricted to a zone several microns behind the actin-rich pseudopod [95], this spatial separation is not entirely consistent with the definition of an invadopodium. Although lamellipodia are rarely observed in 3D matrices because they depend on a planar substrate to extend across, many of the molecular players that are required for lamellopodia are also required for cell migration in 3D environments––e.g. Arp2/3, cofilin, WAVE [92].
Differences between 2D and 3D. Left-hand panels show MTLn3 cell on 2D substrate: note broad lamellipodium, ventral stress fibres and flat cross-section of the cell. Right-hand panels show MTLn3 cell in 3D collagen gel (inset panel shows collagen fibres in white): note absence of stress fibres, more complex organisation of the F-actin at the front of the cell (no longer a planar lamellipodium) and rounded profile of the cell. See also http://london-research-institute.co.uk/research/loc/london/lifch/sahaie/sahaiemoviegallery?view=LRI&source=research_portfolio
Invading A431 squamous cell carcinoma cells do not have a single distinct F-actin protrusion but instead have numerous filopodia [89] (shown in 2D in Fig. 3). It is tempting to speculate that these structures ‘sense’ the surrounding matrix and those that extend in a favourable direction then guide cell movement [96]. However, this will remain a hypothesis until confirmed by experimental studies.
It is also clear that the organization of the acto-myosin contractile machinery can be quite different in more complex environments. Most studies have focused on the regulation of stress fibres, which are prominent in cells cultured on rigid substrates (Fig. 3); however, these structures are much less prominent when cells are in 3D environments [94]. In many cases the contractile machinery is associated with the sub-membranous cortical actin cytoskeleton. Much less is understood about the regulation of this F-actin network; nonetheless it is clear that RhoA and the ROCK kinases are critical for its maintenance. In addition, it has recently been shown that modulation of Dia2 [20] and PDK1 [97, 98] activity can affect cortical actin. High levels of RhoA, RhoC or ROCK activity promote contraction of the cortical actin that is associated with membrane blebbing [99].
Observation of cancer cells moving in 3D environments has suggested that cancer cells can move using series of membrane blebs. Although this type of motility had been observed in vivo in developing fish embryos during the 1970s by Trinkaus and colleagues [100], it received little attention until recently [101]. Strong actin–myosin contraction in one part of a cell may also produce a compressive force that leads to increased hydrostatic pressure and a localized detachment of the plasma membrane from the cortical cytoskeleton which results in bleb protrusion [99]. The cortical actin–myosin network generates a basal level of tension across a cell surface [102]. However, unlike a soap bubble in which surface tension is more or less uniform, local differences in cortical actin–myosin contraction produce variations in tension that affect cell shape. Surface area will increase in regions of localized relaxation whereas contraction will decrease surface area. Consistent with this, ROCK and MLC are localised at the rear of cells moving in this manner [103]. The generation of hydrostatic pressure would require that the contractile machinery be attached to the plasma membrane; in fact, interference with ERM proteins which link acto-myosin cytoskeletal structures with the plasma membrane reduces blebbing-mediated invasion [104]. Although increased intracellular pressure has been observed in blebbing mitotic cells in vitro [105], direct demonstration of the role of hydrostatic pressure in cells moving in 3D environments is problematic, at least in part because techniques that measure force and elasticity such as atomic force microscopy can not easily be used in these environments.
As discussed above, some cancer cells invade with a rounded morphology associated with high levels of Rho–ROCK activity driving cortical acto-myosin contraction; in contrast other cancer cells move with an elongated morphology that does not require Rho–ROCK function [104, 106]. Instead the ROCK related kinases, MRCKα and β function redundantly with ROCK1 and 2 to regulate acto-myosin [47]. ROCK and MLCK have been shown to play distinct but complementary roles in the regulation of MLC phosphorylation, actin structures and motility of cells in 2D tissue culture conditions [107–110]. Elevated MLCK expression has been detected in numerous tumour types [111–114] and cancer cell lines [115–117] suggesting that increased MLCK activity resulting from overexpression or increased calcium transients might act to drive cancer cell motility in vivo, possibly in co-operation with ROCK. ZIPK has recently been shown to be phosphorylated and activated by ROCK [118], and can phosphorylate common substrates including MLC [45], MYPT1 [119] and CPI-17 [68]. These findings suggest that ZIPK could amplify a Rho–ROCK signal or that elevated ZIPK activity might substitute for Rho–ROCK activity, to promote metastasis. However, the exact roles played by MLCK and ZIPK in regulating acto-myosin function in 3D environments remains to be determined.
In addition to the diverse patterns of F-actin organization observed in motile cancer cells [120] (Fig. 3), it is now clear that many cancer cells exhibit significant plasticity in the mechanisms they use to move [104]. This presents a particular challenge when designing inhibitor strategies to block cell movement; for example inhibition of extracellular proteases causes many cancer cells to move with a rounded, blebbing morphology [104, 121]. Constriction of the cortical acto-myosin enables these cells to squeeze through gaps in the surrounding matrix or deform the matrix and thereby invade without the need for protease function [106]. To date, this plasticity has been observed in the experimental context; however, it may also enable cancer cells to overcome the diverse challenges of the metastatic process in human patients. Moving through dense connective tissue, crossing a thin endothelial layer and surviving the shear stresses in the circulation are likely to require different cytoskeletal organizations. Therefore, it may be that a high degree of plasticity in actin organization is particularly favourable during metastasis.
Actin dynamics in living tumours
The use of 3D matrices has highlighted the diversity of motility modes utilized by cancer cells [94]. However, there are always concerns with experimentally generated matrices about how they compare to the matrix surrounding tumours in vivo. Most experimentally generated collagen matrices use pepsin-cleaved collagen I, which lacks the telopeptide of native collagen, and also lacks the cross-linking and higher order organization typical of collagen matrices in tissues [122]. To circumvent these concerns, some researchers have turned to imaging the movement of cancer cells in living tumours [123, 124]. There are a number of methods that have been employed for intravital imaging, including: whole body fluorescence microscopy (most often using confocal or multiphoton microscopes––an example is shown in Fig. 6), implantation of window chambers combined with fluorescence microscopy and whole body bioluminescence [123–126]. These approaches have revealed some surprises: firstly the majority of cancer cells are not motile in vivo even in metastatic tumours [123, 124]; secondly the motile cells frequently move in an ‘amoeboid’ manner that bears similarities to the movement of leukocytes [127] and dispersed Dictyostelium cells [128]. Amoeboid cell motility is fast (>1micron/min) with rapid changes in cell shape and direction (an example is shown in Fig. 6). This leads to cells often having an amorphous appearance. The exact relationship between amoeboid cell motility and the rounded, blebbing associated motility described earlier is not entirely clear. However, there are many similarities including the key role of the cortical F-actin network and the rounded cell morphologies. Gene expression profiling of motile cells collected from metastatic tumours has revealed that these cells coordinatedly up-regulated many of the key actin regulators described above, including cofilin, Arp2/3 complex subunits, N-WASP, LIMK, ROCK1, and RhoA [39]. By combining manipulation of these actin regulators with the in vivo imaging of tumour cells, it is now possible to study the regulation of actin dynamics in tumour cells in situ [123]. This type of approach revealed that inactivation of cofilin by LIMK reduced both tumour cell motility in vivo and metastasis [129], suggesting that actin filament severing by cofilin does indeed have an important in vivo. It will be interesting to examine the role of other regulators of actin polymerization in similar situations.
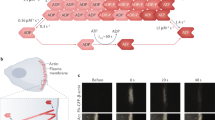
In vivo imaging of amoeboid cell movement. GFP expressing A375 melanoma cells in green, Extra Cellular Matrix in pink, yellow outline shows rapidly moving cell (180s between frames, image 100 × 100 microns). Note rapidly changing morphology and direction of movement (indicated with dashed white line in last panel) and constriction of the cell body at various points (marked with yellow arrowhead). See also http://london-research-institute.co.uk/research/loc/london/lifch/sahaie/sahaiemoviegallery?view=LRI&source=research_portfolio
Live tumour cell imaging will also allow the organization of the cytoskeleton to be analysed and should help to address questions such as; the prevalence of filopodia and invadopodia in motile cells in vivo, and if there are structures analogous to lamellipodia in vivo. By imaging GFP-tagged myosin light chain, the acto-myosin contractile machinery was found to be located around the cortex of motile cancer cells in vivo [106]. Furthermore, the organization of MLC and the motility of these cells was ROCK-dependent [106]. It will be fascinating to extend this type of analysis to regulators of the actin polymerization machinery. The behaviour of cells with increased Rho–ROCK function following stable knockdown of the Smurf1 E3 ubiquitin ligase, which targets RhoA for degradation [130], also has been imaged in vivo. Smurf1 knockdown led to locally increased Rho activity around the cell cortex resulting in a more rounded morphology of motile cells within the tumours and an increased number of cells observed within the vasculature [131]. Taken together, these observations support the notion that high levels of cortical acto-myosin contraction are associated with amoeboid or rounded cancer cell motility and the metastatic process. This could potentially explain the elevated expression levels of many of the molecules involved in the regulation of acto-myosin contraction in metastatic human cancers.
What next?
In this review we have tried to summarize current thinking about regulation of the actin cytoskeleton in invading cancer cells and highlight some areas of current debate. It is clear that there is still much we do not know, but can we speculate what we might hope to learn in next two or three years? Recently the number of known molecules that can promote actin polymerization has increased but many of these have not yet been studied in the context of cancer biology. In fact only the regulators of the Arp2/3 complex and cofilin have been extensively studied in cancer models. It will be fascinating to learn about the role of the various FH proteins and other actin nucleators, such as spire and cordon bleu, in the migration of cancer cells and to determine if they become aberrantly regulated in tumours. Another area of growing interest is diverse range of morphologies or ‘modes of motility’ exhibited by cancer cells; these range from amoeboid, to elongated and collective patterns of invasion. Many human tumours show strand like patterns of invasion with cells often retaining cell-cell adhesions [120]. This adds considerable complexity to the problem of cell invasion; we need to understand how the behaviour of many cells is coordinated so that they invade in one direction, and explore the possibility that distinct cells in the strands have different roles [132]. Greater knowledge of the molecular pathways that determine the mode of motility used by cancer cells and how switching between different actin architectures is regulated will be very beneficial in understanding why and how cancer cells exit primary tumours.
Abbreviations
- 2D:
-
2 Dimensional
- 3D:
-
3 Dimensional
- Arp2/3:
-
Actin related proteins 2 and 3
- CPI-17:
-
PKC-activated protein phosphatase-1 inhibitor
- DAPK:
-
Death-associated protein kinase
- DMPK:
-
Myotonic dystrophy protein kinase
- DRAK:
-
DAP kinase-related apoptosis-inducing protein kinase
- DRF:
-
Diaphanous-related formin
- ECM:
-
Extracellular matrix
- ERM:
-
Ezrin/radixin/moesin
- Ena:
-
Enabled
- F-actin:
-
Filamentous actin
- FH:
-
Formin homology
- GFP:
-
Green fluorescent protein
- ILK:
-
Integrin linked kinase
- LIMK:
-
Lim-domain kinase
- MBS:
-
Myosin-binding subunit
- MLC:
-
Myosin light chain
- MLCK:
-
Myosin light chain kinase
- MRCK:
-
Myotonic dystrophy kinase-related Cdc42–binding kinase
- MYPT:
-
Myosin phosphatase target subunit
- N-WASP:
-
Neural Wiskott-Aldrich syndrome protein
- PAK:
-
p21-Activated kinase
- PDK1:
-
Phosphoinositide dependent protein kinase 1
- PP1M:
-
Myosin protein phosphatase 1
- ROCK:
-
Rho-associated coiled-coil containing kinase
- ROS:
-
Reactive oxygen species
- VASP:
-
Vasodilator-stimulated phosphoprotein
- WASP:
-
Wiskott-Aldrich syndrome protein
- WAVE:
-
WASP family verprolin-homologous protein
- WH2:
-
WASP-homology 2
- ZIPK:
-
Zipper-interacting protein kinase
References
Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465
Rafelski SM, Theriot JA (2004) Crawling toward a unified model of cell motility: spatial and temporal regulation of actin dynamics. Annu Rev Biochem 73:209–239
Zigmond SH, Gerald PS (2004) Beginning and ending an actin filament: control at the barbed end. In: current topics in developmental biology, vol 63. Academic Press, pp 145–188
Goode BL, Eck MJ (2007) Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76:593–627
Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB (2003) Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol 19:541–564
Pollard TD (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct 36:451–477
Wang W, Eddy R, Condeelis J (2007) The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer 7:429–440
Jaffe AB, Hall A (2005) Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 21:247–269
Logan MR, Mandato CA (2006) Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell 98:377–388
Wear MA, Cooper JA (2004) Capping protein: new insights into mechanism and regulation. Trends Biochem Sci 29:418–428
Yarmola EG, Bubb MR (2006) Profilin: emerging concepts and lingering misconceptions. Trends Biochem Sci 31:197–205
Takenawa T, Suetsugu S (2007) The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8:37–48
Suetsugu S, Miki H, Takenawa T (1998) The essential role of profilin in the assembly of actin for microspike formation. Embo J 17:6516–6526
Dominguez R (2007) The beta-thymosin/WH2 fold: multifunctionality and structure. Ann N Y Acad Sci 1112:86–94
Carlier MF, Hertzog M, Didry D, Renault L, Cantrelle FX, van Heijenoort C, Knossow M, Guittet E (2007) Structure, function, and evolution of the beta-thymosin/WH2 (WASP-Homology2) actin-binding module. Ann N Y Acad Sci 1112:67–75
Scita G, Confalonieri S, Lappalainen P, Suetsugu S (2008) IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol 18:52–60
Buday L, Downward J (2007) Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta 1775:263–273
Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS (2003) Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 (mDia2) as an effector for Cdc42. Curr Biol 13:534–545
Wallar BJ, Deward AD, Resau JH, Alberts AS (2007) RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res 313:560–571
Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS (2007) Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol 17:579–591
Pellegrin S, Mellor H (2005) The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol 15:129–133
Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S (1997) p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. Embo J 16:3044–3056
Zigmond SH (2004) Formin-induced nucleation of actin filaments. Curr Opin Cell Biol 16:99–105
Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA (2007) Src phosphorylation of cortactin enhances actin assembly. Proc Natl Acad Sci U S A 104:11933–11938
Scott RW, Olson MF (2007) LIM kinases: function, regulation and association with human disease. J Mol Med 85:555–568
Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG (2003) RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol 162:1267–1279
Takeya R, Taniguchi K, Narumiya S, Sumimoto H (2008) The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. Embo J 27:618–628
Schirenbeck A, Arasada R, Bretschneider T, Schleicher M, Faix J (2005) Formins and VASPs may co-operate in the formation of filopodia. Biochem Soc Trans 33:1256–1259
Chhabra ES, Higgs HN (2007) The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol 9:1110–1121
Adams JC (2004) Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol 16:590–596
Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174:863–875
O’Connell CB, Tyska MJ, Mooseker MS (2007) Myosin at work: motor adaptations for a variety of cellular functions. Biochim Biophys Acta 1773:615–630
Gunning PW, Schevzov G, Kee AJ, Hardeman EC (2005) Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 15:333–341
Ichetovkin I, Grant W, Condeelis J (2002) Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol 12:79–84
DesMarais V, Macaluso F, Condeelis J, Bailly M (2004) Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci 117:3499–3510
van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, Yip SC, Backer JM, Eddy RJ, Condeelis JS (2007) EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol 179:1247–1259
El-Sibai M, Nalbant P, Pang H, Flinn RJ, Sarmiento C, Macaluso F, Cammer M, Condeelis JS, Hahn KM, Backer JM (2007) Cdc42 is required for EGF-stimulated protrusion and motility in MTLn3 carcinoma cells. J Cell Sci 120:3465–3474
Yip SC, El-Sibai M, Coniglio SJ, Mouneimne G, Eddy RJ, Drees BE, Neilsen PO, Goswami S, Symons M, Condeelis JS, Backer JM (2007) The distinct roles of Ras and Rac in PI 3-kinase-dependent protrusion during EGF-stimulated cell migration. J Cell Sci 120:3138–3146
Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS (2004) Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 64:8585–8594
Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TEB (2006) Filopodia Formation in the Absence of Functional WAVE- and Arp2/3-Complexes. Mol Biol Cell 17:2581–2591
Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB (2002) Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109:509–521
Vale RD, Milligan RA (2000) The way things move: looking under the hood of molecular motor proteins. Science 288:88–95
Somlyo AP, Somlyo AV (2000) Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J Physiol 522(Pt 2):177–185
Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho- kinase). J Biol Chem 271:20246–20249
Murata-Hori M, Suizu F, Iwasaki T, Kikuchi A, Hosoya H (1999) ZIP kinase identified as a novel myosin regulatory light chain kinase in HeLa cells. FEBS Lett 451:81–84
Leung T, Chen XQ, Tan I, Manser E, Lim L (1998) Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol 18:130–140
Wilkinson S, Paterson HF, Marshall CJ (2005) Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat Cell Biol 7:255–261
Wilson DP, Sutherland C, Borman MA, Deng JT, Macdonald JA, Walsh MP (2005) Integrin-linked kinase is responsible for Ca2+-independent myosin diphosphorylation and contraction of vascular smooth muscle. Biochem J 392:641–648
Cohen O, Feinstein E, Kimchi A (1997) DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. Embo J 16:998–1008
Kawai T, Nomura F, Hoshino K, Copeland NG, Gilbert DJ, Jenkins NA, Akira S (1999) Death-associated protein kinase 2 is a new calcium/calmodulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene 18:3471–3480
Sanjo H, Kawai T, Akira S (1998) DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem 273:29066–29071
Ramos E, Wysolmerski RB, Masaracchia RA (1997) Myosin phosphorylation by human cdc42-dependent S6/H4 kinase/gammaPAK from placenta and lymphoid cells. Recept Signal Transduct 7:99–110
Zeng Q, Lagunoff D, Masaracchia R, Goeckeler Z, Cote G, Wysolmerski R (2000) Endothelial cell retraction is induced by PAK2 monophosphorylation of myosin II. J Cell Sci 113(Pt 3):471–482
Guerriero V Jr., Russo MA, Olson NJ, Putkey JA, Means AR (1986) Domain organization of chicken gizzard myosin light chain kinase deduced from a cloned cDNA. Biochemistry 25:8372–8381
Ito M, Nakano T, Erdodi F, Hartshorne DJ (2004) Myosin phosphatase: structure, regulation and function. Mol Cell Biochem 259:197–209
Okamoto R, Kato T, Mizoguchi A, Takahashi N, Nakakuki T, Mizutani H, Isaka N, Imanaka-Yoshida K, Kaibuchi K, Lu Z, Mabuchi K, Tao T, Hartshorne DJ, Nakano T, Ito M (2006) Characterization and function of MYPT2, a target subunit of myosin phosphatase in heart. Cell Signal 18:1408–1416
Feng J, Ito M, Ichikawa K, Isaka N, Nishikawa M, Hartshorne DJ, Nakano T (1999) Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem 274:37385–37390
Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R (2004) Structural basis of protein phosphatase 1 regulation. Nature 429:780–784
Tan I, Ng CH, Lim L, Leung T (2001) Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J Biol Chem 276:21209–21216
Muranyi A, MacDonald JA, Deng JT, Wilson DP, Haystead TA, Walsh MP, Erdodi F, Kiss E, Wu Y, Hartshorne DJ (2002) Phosphorylation of the myosin phosphatase target subunit by integrin-linked kinase. Biochem J 366:211–216
Kiss E, Muranyi A, Csortos C, Gergely P, Ito M, Hartshorne DJ, Erdodi F (2002) Integrin-linked kinase phosphorylates the myosin phosphatase target subunit at the inhibitory site in platelet cytoskeleton. Biochem J 365:79–87
Borman MA, MacDonald JA, Muranyi A, Hartshorne DJ, Haystead TAJ (2002) Smooth muscle myosin phosphatase-associated kinase induces Ca2+ sensitization via myosin phosphatase inhibition. J Biol Chem 277:23441–23446
Muranyi A, Zhang R, Liu F, Hirano K, Ito M, Epstein HF, Hartshorne DJ (2001) Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett 493:80–84
Velasco G, Armstrong C, Morrice N, Frame S, Cohen P (2002) Phosphorylation of the regulatory subunit of smooth muscle protein phosphatase 1M at Thr850 induces its dissociation from myosin. FEBS Lett 527:101–104
Li L, Eto M, Lee MR, Morita F, Yazawa M, Kitazawa T (1998) Possible involvement of the novel CPI-17 protein in protein kinase C signal transduction of rabbit arterial smooth muscle. J Physiol 508(Pt 3):871–881
Eto M, Kitazawa T, Matsuzawa F, Aikawa S, Kirkbride JA, Isozumi N, Nishimura Y, Brautigan DL, Ohki SY (2007) Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor. Structure 15:1591–1602
Koyama M, Ito M, Feng J, Seko T, Shiraki K, Takase K, Hartshorne DJ, Nakano T (2000) Phosphorylation of CPI-17, an inhibitory phosphoprotein of smooth muscle myosin phosphatase, by Rho-kinase. FEBS Lett 475:197–200
MacDonald JA, Eto M, Borman MA, Brautigan DL, Haystead TA (2001) Dual Ser and Thr phosphorylation of CPI-17, an inhibitor of myosin phosphatase, by MYPT-associated kinase. FEBS Lett 493:91–94
Deng JT, Sutherland C, Brautigan DL, Eto M, Walsh MP (2002) Phosphorylation of the myosin phosphatase inhibitors, CPI-17 and PHI-1, by integrin-linked kinase. Biochem J 367:517–524
Eto M, Karginov A, Brautigan DL (1999) A novel phosphoprotein inhibitor of protein type-1 phosphatase holoenzymes. Biochemistry 38:16952–16957
Erdodi F, Kiss E, Walsh MP, Stefansson B, Deng JT, Eto M, Brautigan DL, Hartshorne DJ (2003) Phosphorylation of protein phosphatase type-1 inhibitory proteins by integrin-linked kinase and cyclic nucleotide-dependent protein kinases. Biochem Biophys Res Commun 306:382–387
Jin H, Sperka T, Herrlich P, Morrison H (2006) Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 442:576–579
Somlyo AP, Somlyo AV (2003) Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev 83:1325–1358
Garrett SC, Varney KM, Weber DJ, Bresnick AR (2006) S100A4, a mediator of metastasis. J Biol Chem 281:677–680
Kriajevska MV, Cardenas MN, Grigorian MS, Ambartsumian NS, Georgiev GP, Lukanidin EM (1994) Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J Biol Chem 269:19679–19682
Li ZH, Bresnick AR (2006) The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res 66:5173–5180
Watanabe Y, Usada N, Minami H, Morita T, Tsugane S, Ishikawa R, Kohama K, Tomida Y, Hidaka H (1993) Calvasculin, as a factor affecting the microfilament assemblies in rat fibroblasts transfected by src gene. FEBS Lett 324:51–55
Takenaga K, Nakamura Y, Sakiyama S, Hasegawa Y, Sato K, Endo H (1994) Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J Cell Biol 124:757–768
Stehn JR, Schevzov G, O’Neill GM, Gunning PW (2006) Specialisation of the tropomyosin composition of actin filaments provides new potential targets for chemotherapy. Curr Cancer Drug Targets 6:245–256
Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJC, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP (2003) Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell 14:1002–1016
Fanning AS, Wolenski JS, Mooseker MS, Izant JG (1994) Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton 29:29–45
Adami R, Cintio O, Trombetta G, Choquet D, Grazi E (2003) On the stiffness of the natural actin filament decorated with alexa fluor tropomyosin. Biophys Chem 104:469–476
Ono S, Ono K (2002) Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol 156:1065–1076
Ishikawa R, Yamashiro S, Matsumura F (1989) Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem 264:7490–7497
Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G (2004) Two distinct actin networks drive the protrusion of migrating cells. Science 305:1782–1786
DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE (2002) Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci 115:4649–4660
Soderling SH, Scott JD (2006) WAVE signalling: from biochemistry to biology. Biochem Soc Trans 34:73–76
Nimnual AS, Taylor LJ, Bar-Sagi D (2003) Redox-dependent downregulation of Rho by Rac. Nat Cell Biol 5:236–241
Hooper S, Marshall JF, Sahai E (2006) Tumor cell migration in three dimensions. Meth Enzymol 406:625–643
Buccione R, Orth JD, McNiven MA (2004) Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol 5:647–657
Linder S (2007) The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol 17:107–117
Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J (2005) Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol 168:441–452
Yamaguchi H, Pixley F, Condeelis J (2006) Invadopodia and podosomes in tumor invasion. Eur J Cell Biol 85:213–218
Yamada KM, Cukierman E (2007) Modeling tissue morphogenesis and cancer in 3D. Cell 130:601–610
Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P (2007) Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol 9:893–904
Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ (2005) Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol 170:619–626
Zaru R, Mollahan P, Watts C (2008) 3-Phosphoinositide-dependent kinase 1 deficiency perturbs toll-like receptor signaling events and actin cytoskeleton dynamics in dendritic cells. J Biol Chem 283:929–939
Pinner S, Sahai E (2008) PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat Cell Biol 10:127–137
Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ (2005) Non-equilibration of hydrostatic pressure in blebbing cells. Nature 435:365–369
Trinkaus JP (1973) Surface activity and locomotion of Fundulus deep cells during blastula and gastrula stages. Dev Biol 30:69–103
Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, Solnica-Krezel L, Heisenberg CP, Raz E (2006) Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell 11:613–627
Lecuit T, Lenne P-F (2007) Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 8:633–644
Pinner S, Sahai E (2008) PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat Cell Biol 10:127–137
Sahai E, Marshall CJ (2003) Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol 5:711–719
Charras GT, Coughlin M, Mitchison TJ, Mahadevan L (2007) Life and times of a cellular bleb. Biophys J 94:1836–1853
Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E (2006) ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol 16:1515–1523
Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F (2000) Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol 150:797–806
Totsukawa G, Wu Y, Sasaki Y, Hartshorne DJ, Yamakita Y, Yamashiro S, Matsumura F (2004) Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J Cell Biol 164:427–439
Chen BH, Tzen JT, Bresnick AR, Chen HC (2002) Roles of Rho-associated kinase and myosin light chain kinase in morphological and migratory defects of focal adhesion kinase-null cells. J Biol Chem 277:33857–33863
Niggli V, Schmid M, Nievergelt A (2006) Differential roles of Rho-kinase and myosin light chain kinase in regulating shape, adhesion, and migration of HT1080 fibrosarcoma cells. Biochem Biophys Res Commun 343:602–608
Lee WS, Seo G, Shin HJ, Yun SH, Yun H, Choi N, Lee J, Son D, Cho J, Kim J, Cho YB, Chun HK, Lee WY (2008) Identification of differentially expressed genes in microsatellite stable HNPCC and sporadic colon cancer. J Surg Res 144:29–35
Minamiya Y, Nakagawa T, Saito H, Matsuzaki I, Taguchi K, Ito M, Ogawa J (2005) Increased expression of myosin light chain kinase mRNA is related to metastasis in non-small cell lung cancer. Tumour Biol 26:153–157
Ye LH, Wu LY, Guo W, Ma HT, Zhang XD (2006) [Screening of a sub-clone of human breast cancer cells with high metastasis potential]. Zhonghua Yi Xue Za Zhi 86:61–65
Kucharczak J, Pannequin J, Camby I, Decaestecker C, Kiss R, Martinez J (2001) Gastrin induces over-expression of genes involved in human U373 glioblastoma cell migration. Oncogene 20:7021–7028
Tohtong R, Phattarasakul K, Jiraviriyakul A, Sutthiphongchai T (2003) Dependence of metastatic cancer cell invasion on MLCK-catalyzed phosphorylation of myosin regulatory light chain. Prostate Cancer Prostatic Dis 6:212–216
Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T (2002) Myosin light chain kinase inhibitors can block invasion and adhesion of human pancreatic cancer cell lines. Pancreas 24:34–41
Nguyen Q-D, Faivre S, Bruyneel E, Rivat C, Seto M, Endo T, Mareel M, Emami S, Gespach C (2002) RhoA- and RhoD-dependent regulatory switch of G{alpha} subunit signaling by PAR-1 receptors in cellular invasion. FASEB J 16:565–576
Hagerty L, Weitzel DH, Chambers J, Fortner CN, Brush MH, Loiselle D, Hosoya H, Haystead TA (2007) ROCK1 phosphorylates and activates zipper-interacting protein kinase. J Biol Chem 282:4884–4893
MacDonald JA, Borman MA, Muranyi A, Somlyo AV, Hartshorne DJ, Haystead TAJ (2001) Identification of the endogenous smooth muscle myosin phosphatase-associated kinase. Proc Natl Acad Sci 98:2419–2424
Sahai E (2005) Mechanisms of cancer cell invasion. Curr Opin Genet Dev 15:87–96
Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker E-B, Friedl P (2003) Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 160:267–277
Woodley DT, Yamauchi M, Wynn KC, Mechanic G, Briggaman RA (1991) Collagen telopeptides (Cross-Linking Sites) play a role in collagen gel lattice contraction. J Investig Dermatol 97:580–585
Sahai E (2007) Illuminating the metastatic process. Nat Rev Cancer 7:737–749
Condeelis J, Segall JE (2003) Intravital imaging of cell movement in tumours. Nat Rev Cancer 3:921–930
Makale M (2007) Intravital imaging and cell invasion. Meth Enzymol 426:375–401
MacDonald IC, Groom AC, Chambers AF (2002) Cancer spread and micrometastasis development: quantitative approaches for in vivo models. Bioessays 24:885–893
Friedl P, Wolf K (2003) Proteolytic and non-proteolytic migration of tumour cells and leucocytes. Biochem Soc Symp 70:277–285
Friedl P, Borgmann S, Brocker EB (2001) Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J Leukoc Biol 70:491–509
Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS (2006) The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 173:395–404
Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL (2003) Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302:1775–1779
Sahai E, Garcia-Medina R, Pouyssegur J, Vial E (2007) Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol 176:35–42
Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E (2007) Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 9:1392–1400
Acknowledgements
This work was supported by Cancer Research UK (Erik Sahai and Michael F. Olson) and by the National Institutes of Health (Michael F. Olson; R01 CA030721).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Olson, M.F., Sahai, E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis 26, 273–287 (2009). https://doi.org/10.1007/s10585-008-9174-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-008-9174-2