Published online Sep 28, 2010. doi: 10.3748/wjg.v16.i36.4532
Revised: May 5, 2010
Accepted: May 12, 2010
Published online: September 28, 2010
AIM: To investigate the correlations between self-reported symptoms of irritable bowel syndrome (IBS) and the gastrointestinal (GI) microbiota composition.
METHODS: Fecal samples were collected from a total of 44 subjects diagnosed with IBS. Their symptoms were monitored with a validated inflammatory bowel disease questionnaire adjusted for IBS patients. Thirteen quantitative real-time polymerase chain reaction assays were applied to evaluate the GI microbiota composition. Eubacteria and GI bacterial genera (Bifidobacterium, Lactobacillus and Veillonella), groups (Clostridium coccoides/Eubacterium rectale, Desulfovibrio desulfuricans) and distinct bacterial phylotypes [closest 16S rDNA sequence resemblance to species Bifidobacterium catenulatum, Clostridium cocleatum, Collinsella aerofaciens (C. aerofaciens), Coprococcus eutactus (C. eutactus), Ruminococcus torques and Streptococcus bovis] with a suspected association with IBS were quantified. Correlations between quantities or presence/absence data of selected bacterial groups or phylotypes and various IBS-related symptoms were investigated.
RESULTS: Associations were observed between subjects’ self-reported symptoms and the presence or quantities of certain GI bacteria. A Ruminococcus torques (R. torques)-like (94% similarity in 16S rRNA gene sequence) phylotype was associated with severity of bowel symptoms. Furthermore, among IBS subjects with R. torques 94% detected, the amounts of C. cocleatum 88%, C. aerofaciens-like and C. eutactus 97% phylotypes were significantly reduced. Interesting observations were also made concerning the effect of a subject’s weight on GI microbiota with regard to C. aerofaciens-like phylotype, Bifidobacterium spp. and Lactobacillus spp.
CONCLUSION: Bacteria seemingly affecting the symptom scores are unlikely to be the underlying cause or cure of IBS, but they may serve as biomarkers of the condition.
- Citation: Malinen E, Krogius-Kurikka L, Lyra A, Nikkilä J, Jääskeläinen A, Rinttilä T, Vilpponen-Salmela T, Wright AJV, Palva A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol 2010; 16(36): 4532-4540
- URL: https://www.wjgnet.com/1007-9327/full/v16/i36/4532.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i36.4532
Irritable bowel syndrome (IBS) is a common functional bowel disorder, with an estimated worldwide prevalence of 10%-20% among adults and adolescents. IBS is characterized by pain or discomfort, distorted bowel habits and altered stool characteristics[1]. Although the prognosis of IBS is good, the syndrome results in a reduced quality of life. Subjects with IBS report significantly more comorbidities, including dyspepsia, asthma and head- and backache, as well as anxiety, depression and insomnia, than the general population[2]. The exact etiology of IBS is likely to be multifactorial; moreover, patients diagnosed with the disorder may well be experiencing bowel symptoms due to different causes.
Much attention has recently been focused on the impact of gastrointestinal (GI) microbiota on this disorder. Changes observed in the fecal microbiota composition[3-5], high incidence of IBS after GI infections[6], alterations seen in IBS patients’ GI immune systems[7], as well as ability of probiotics to alleviate the symptoms of IBS[8-10], all suggest that microbes play a key role in IBS. However, a genetic basis for IBS has also been presented. In twin studies, a greater likelihood for a twin to develop IBS if the other sibling already had the disorder was observed among monozygotic twins compared to dizygotic twins[11]. Upregulation of genes involved in mucin production has been described to take place in IBS patients[12]. Downregulation of protease-activated receptor 1 expression and upregulation of protease-activated receptor 2 ligand mast cell tryptase in diarrhea-predominant IBS (IBS-D) are involved in visceral hypersensitivity; a change in the expression ratio of these two protease-activated receptors appears to take place in the context of IBS-D[13]. Similarly to ulcerative colitis, colonic mucosal 5-HT (serotonin) concentrations are significantly lowered in IBS patients compared with the levels observed in healthy controls, suggesting the existence of a shared mechanism underlying the symptoms[14].
In this study, we examined whether the presence or absence of certain microbes previously linked to either IBS or healthy controls’ microbiota correlated with the symptoms experienced by IBS patients. Our results suggest that a connection between IBS-related microbiota and severity of self-reported symptoms exists.
Subjects fulfilling the Rome I criteria for IBS[15] were recruited from the district of Kuopio in Eastern Finland by the Kuopio University Hospital and Harjula Hospital during years 2004-2005. The participants (n = 44; 11 men, 33 women) were 20-72 years old and their general condition was confirmed as good by medical experts (see Table 1 for subject characteristics). Exclusion criteria for participation included presence of organic GI diseases, inadequately treated hypertension or pharmaceutically treated diabetes. Use of statins, pharmaceutically treated hypertension or coronary artery disease were not considered exclusion criteria if medication had been used for at least six months prior to the study with no changes in dosage.
| Characteristic | mean ± SD | Median (range) |
| Age (yr) | 48.4 ± 11.9 | 49.0 (20-72) |
| Body mass index (kg/m2) | 26.3 ± 5.1 | 26.0 (19.4-45.6) |
| Systolic blood pressure (mmHg) | 129.2 ± 19.1 | 129.5 (95-175) |
| Diastolic blood pressure (mmHg) | 83.0 ± 10.2 | 83.5 (61-100) |
| Hemoglobin (g/L) | 139.0 ± 12.0 | 139.0 (108-165) |
| B-GHba1c (%) | 5.6 ± 0.38 | 5.6 (4.8-6.4) |
| Total cholesterol (mmol/L) | 5.1 ± 0.73 | 5.0 (3.8-7.3) |
| Sedimentation rate (mm/h) | 7.4 ± 7.1 | 5.0 (2-43) |
Participants were subjected to a standard medical examination and laboratory tests, including blood cell counts (B-Leuk, B-Trom, B-Eryt), hemoglobin (B-Hb), hematocrit (B-Hcr), erythrocyte mean cell volume (E-MCV), mean cell hemoglobin (E-MCH) and mean corpuscular hemoglobin concentration (E-MCHC), erythrocyte sedimentation rate, glycosylated hemoglobin (B-GHbAlc) and blood lipids (fS-Chol, fS-Chol-HDL, S-Trigly). Lactose absorption was ensured with a DNA test for a mutation in the lactase gene. The participants were also tested for the presence of IgA antibodies against gliadin, followed by a duodenal biopsy if celiac disease was suspected. Subjects over 45 years were examined for the presence of carcinoembryonic antigen (CEA). In addition, presence of occult blood in the feces was evaluated.
Participants filled in a questionnaire regarding their quality of life and symptoms. The survey was based on an internationally approved and validated questionnaire for inflammatory bowel disease questionnaire (IBDQ)[16]. The questions in the IBDQ were adapted to suit IBS patients. The query contained 30 questions clustered into four groups, comprising “bowel symptoms”, “systemic symptoms”, “social function” and “emotional function”. The seven alternative answers for each question were assigned numerical values from 1 to 7 (1 = no problem at all, 7 = very severe problem). Two questions regarding the form of feces and the frequency of defecation were treated as separate variables. The participants also answered two questions about prior antibiotic treatments and GI infections.
Each subject gave a fecal sample for bacteriological studies. The samples were stored at -80°C prior to analysis. Bacterial DNA was isolated from 1 g of fecal material by removing the undigested particles from the fecal material with three rounds of low-speed centrifugation, collection of the bacterial cells with high-speed centrifugation, enzymatic and mechanical cell lysis and DNA extraction and precipitation[17]. DNA concentrations were measured with the multilabel plate reader Victor3TM (PerkinElmer Life and Analytical Sciences, Boston, MA, USA). With the method, the average yield of DNA was 342 μg/g of fecal material (median: 290 μg/g, SD: 167 μg/g). Quantitative real-time polymerase chain reaction (qPCR) was used to assess the amounts of selected GI bacteria or bacterial phylotypes (Table 2) in the fecal samples.
| Target bacterial group/phylotype | Positive control strain or clone | MgCl2 (mmol/L) | Detection (°C) | Annealing (°C) | Primer sequences (F: forward, R: reverse) |
| Bifidobacterium catenulatum/Pseudocatenulatum-like[4] | AM277302 | 3 | 87 | 68 | F: 5'-ACTCCTCGCATGGGGTGTC-3' |
| R: 5'-CCGAAGGCTTGCTCCCGAT-3' | |||||
| Bifidobacterium spp.[30] | Bifidobacterium longum | 3 | 85 | 58 | F: 5'-TCGCGTC(C/T)GGTGTGAAAG-3' |
| DSM20219 | R: 5'-CCACATCCAGC(A/G)TCCAC-3' | ||||
| Clostridium coccoides/Eubacterium rectale-group[30] | Ruminococcus productus | 4 | 85 | 55 | F: 5'-CGGTACCTGACTAAGAAG-3' |
| R: 5'-AGTTT(C/T)ATTCTTGCGAAC-3' | |||||
| DSM2950 | |||||
| Clostridium cocleatum 88%[4] | AM275477 | 4 | 80 | 60 | F: 5'-AATACATAAGTAACCTGGCRTC-3' |
| R: 5'-CGTAGCACTTTTCATATAGAGTT-3' | |||||
| Collinsella aerofaciens-like[4] | AM276364 | 4 | 89 | 67 | F: 5'-CCCGACGGGAGGGGAT-3' |
| R: 5'-CTTCTGCAGGTACAGTCTTGAC-3' | |||||
| Coprococcus eutactus 97%[4] | AM278899 | 2 | 83 | 63 | F: 5'-AGCTTGCTCCGGCYGATTTA-3' |
| R: 5'-CGGTTTTACCAGTCGTTTCCAA-3' | |||||
| Desulfovibrio desulfuricans-group[30] | Desulfovibrio desulfuricans | 4 | 85 | 58 | F: 5'-GGTACCTTCAAAGGAAGCAC-3' |
| ATCC7757 | R: 5'-GGGATTTCACCCCTGACTTA-3' | ||||
| Eubacterial 16S[31] | Bifidobacterium longum | 3 | 80 | 50 | F: 5'-TCCTACGGGAGGCAGCAGT-3' |
| DSM20219 | R: 5'-GGACTACCAGGGTATCTAATCCTGTT-3' | ||||
| Lactobacillus-group[32,33] | Lactobacillus acidophilus | 2 | 85 | 58 | F: 5'-AGCAGTAGGGAATCTTCCA-3' |
| ATCC4356 | R: 5'-CACCGCTACACATGGAG-3' | ||||
| Ruminococcus torques 91%[4] | AM276624 | 5 | 82 | 62 | F: 5'-TGCTTAACTGATCTTCTTCGGA-3' |
| R: 5'-CGGTATTAGCAGTCATTTCTG-3' | |||||
| Ruminococcus torques 94%[4] | AM277929 | 2 | 85 | 65 | F: 5'-AATCTTCGGAGGAAGAGGACA-3' |
| R: 5'-ACACTACACCATGCGGTCCT-3' | |||||
| Streptococcus bovis-like[4] | AM276479 | 5 | 80 | 60 | F: 5'-TTAGCTTGCTAAAGTTGGAA-3' |
| R: 5'-ATCTACTAGTGAAGCAATTGCT-3' | |||||
| Veillonella spp.[30] | Veillonella parvula ATCC10790 | 3 | 85 | 62 | F: 5'-A(C/T)CAACCTGCCCTTCAGA-3' |
| R: 5'-CGTCCCGATTAACAGAGCTT-3' |
In total, 13 qPCR assays were performed to analyze the GI microbiota in fecal samples (Table 2). The applied assays targeted quantitatively IBS-associated human GI bacteria (Lactobacillus spp., Clostridium coccoides/Eubacterium rectale-group, Veillonella spp. and Bifidobacterium spp.)[5] and bacterial phylotypes [Collinsella aerofaciens-like, Clostridium cocleatum 88%, Coprococcus eutactus 97%, Ruminococcus torques 91% and Ruminococcus torques (R. torques) 94%][4,18] or bacteria associated with IBS in semi-quantitative sequence data analyses (Bifidobacterium catenulatum/Pseudocatenulatum-like)[4] or with intestinal disturbances according to the literature (Desulfovibrio desulfuricans-group)[19]. The iCycler iQ Real-Time Detection System (Bio-Rad, Hercules, CA, USA) in conjunction with the iCycler Optical System Interface software (version 2.3; Bio-Rad) were used to analyze the samples as described previously[4,5]. Two technical replicates were used for the samples and standard reactions. Depending on the assay, 0.5 or 50 ng of fecal DNA was applied in the reactions.
Basic statistical analysis of the data was performed using the SPSS program, version 14.0 (SPSS Inc., Chicago, IL, USA). R program, version 2.8.0[20] and the package Rcmdr, version 1.4-10[21] were used to perform the principal component analysis (PCA), to describe the categorical data and to statistically test this data. Linear models were used to describe the relationship between variables and were applied to each bacterial genus and phylotype quantified.
The health-related quality of life questionnaire yielded ordinal data; thus, non-parametric statistical methods were used for analysis. However, PCA analysis was performed for the questionnaire data. Results are presented as medians and interquartile ranges. The χ2 test was applied to compare nominal data, and the Mann-Whitney U test to compare continuous data when two patient groups were analyzed.
Logarithmic transformation was performed on the bacterial data prior to further analyses. Within the data, undetected abundances possibly caused by technical limitations instead of the absence of the target phylotype were imputed with the mean values obtained from the qPCR runs with the same primer applied to water. If these also were undetected for a certain assay, the minimum of all detected water runs was used. Bacterial qPCR data were also inspected to ascertain the presence or absence of certain phylotypes in the patient samples. Binary data were then used to evaluate whether the phylotypes had any relationship with various traits of the study subjects.
The study protocol was approved by the Kuopio University Hospital Ethical Committee. Participation in the study was voluntary, and patients were allowed to withdraw at any point without giving an explanation.
Characteristics of the IBS patients are listed in Table 1. In general, the clinical studies revealed no major issues regarding the health status of participants. However, of the 44 subjects studied, 7/11 men and 15/33 women had a body mass index (BMI) value above 25, which is considered borderline between normal weight and slightly overweight[22]. Presence of Helicobacter had been confirmed previously in 8 patients; interestingly, in some cases, treatment of the infection had remained incomplete (data not shown). Helicobacter infection or the way it had been treated was not, however, reflected in the symptoms (data not shown). Origin of IBS as a result of GI inflammation was not given support by this study, as only one subject recalled suffering from gastroenteritis but could not remember whether the onset of IBS occurred before or after the ailment.
The modified IBDQ symptom questionnaire consisted of 28 questions divided into the four categories of bowel symptoms, systemic symptoms, social function and emotional function (Table 3). High median values along with a narrow interquartile range can be considered characteristic of the questions central for ascertaining symptoms of IBS (Table 3). Correlations between the four categories were all significant (Table 4).
| Symptom groups | Median (interquartile range) | Minimum value | Maximum value |
| Bowel symptoms | |||
| Increased frequency of defecation | 3 (2) | 1 | 6 |
| Abdominal cramps | 3 (1) | 1 | 5 |
| Passing gas | 4 (2) | 1 | 7 |
| Abdominal bloating | 5 (2) | 1 | 7 |
| Feeling a need to defecate | 3 (2) | 1 | 7 |
| Soiling | 1 (1.25) | 1 | 5 |
| Stomach pain | 3 (1) | 1 | 6 |
| Systemic symptoms | |||
| Tired and worn out | 3.5 (1) | 1 | 6 |
| Nausea | 3 (2) | 1 | 6 |
| Generally unwell | 3 (1.25) | 1 | 5 |
| Sleep disturbance | 4 (2) | 1 | 7 |
| Weight problems | 3 (2.25) | 1 | 7 |
| Social function | |||
| Work/school activities | 1 (0) | 1 | 7 |
| Cancel social engagements | 1 (1) | 1 | 7 |
| Leisure/sports activity | 3 (3) | 1 | 7 |
| Avoid events lacking toilet | 1 (1) | 1 | 4 |
| Sexual activity | 2 (2) | 1 | 7 |
| Emotional function | |||
| Frustrated/impatient | 3 (1) | 1 | 6 |
| Energy | 3 (2) | 2 | 6 |
| Fear of not finding a toilet | 2 (2) | 1 | 4 |
| Depressed/discouraged | 3 (1) | 1 | 5 |
| Worried about illness | 3 (1.25) | 1 | 7 |
| Relaxed/free from worries | 5 (1) | 2 | 7 |
| Tearful or upset | 2 (1.25) | 1 | 5 |
| Angry | 3 (2) | 1 | 7 |
| Irritable | 3 (2) | 1 | 5 |
| Lack of understanding by others | 2 (1) | 1 | 6 |
| Satisfaction with personal life | 4 (2) | 1 | 6 |
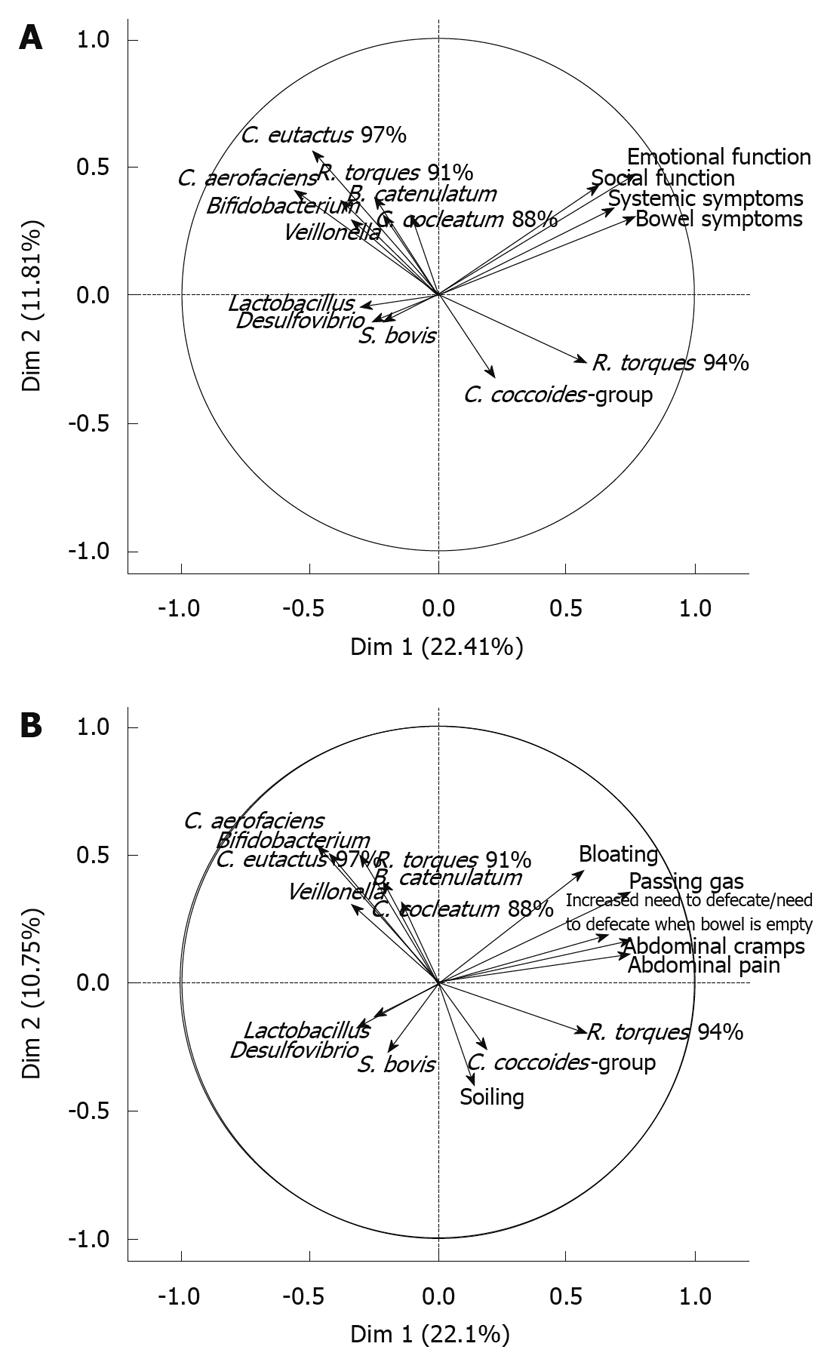
Abundance and prevalence of the 13 qPCR target bacteria or phylotypes in patient samples are shown in Tables 5 and 6, respectively. An association between R. torques 94% phylotype and symptom scores (emotional function, social function, systemic symptoms, bowel symptoms) was observed in a PCA visualization of the results as they correlated significantly with the same dimension, whereas a weaker negative association was observed for Coprococcus eutactus (C. eutactus) 97%, Bifidobacterium spp., Veillonella spp. and Desulfovibrio desulfuricans (D. desulfuricans)-group and the symptom scores (Figure 1A). When the bowel symptoms (bloating, passing gas, increased need to defecate or need to defecate when bowel is empty, abdominal cramps, abdominal pain, soiling) were analyzed in a PCA, a similar effect was observed: R. torques 94% and all bowel symptoms except soiling correlated with the same dimension, whereas Collinsella aerofaciens (C. aerofaciens)-like, C. eutactus 97%, Veillonella spp., Bifidobacterium spp., and Lactobacillus spp. were negatively associated (Figure 1B).
| qPCR assay | All (n = 44) | BMI < 251 (n = 17) | BMI > 25 (n = 22) | P2 |
| Bifidobacterium catenulatum/Pseudocatenulatum-like | 4.8 (1.6)3 | 5.4 (0.7) | 4.3 (1.8) | 0.366 |
| Bifidobacterium spp. | 5.8 (1.0) | 5.5 (1.0) | 6.2 (0.9) | 0.009 |
| Clostridium coccoides/Eubacterium rectale-group | 7.3 (0.2) | 7.3 (0.1) | 7.3 (0.2) | 0.630 |
| Clostridium cocleatum 88% | 5.6 (1.1) | 5.5 (1.4) | 5.6 (1.0) | 0.483 |
| Collinsella aerofaciens-like | 5.6 (1.1) | 5.3 (1.2) | 5.8 (0.9) | 0.294 |
| Coprococcus eutactus 97% | 5.3 (1.5) | 5.5 (1.7) | 5.4 (1.3) | 0.639 |
| Desulfovibrio desulfuricans-group | 3.9 (0.8) | 3.8 (0.6) | 4.0 (0.9) | 0.403 |
| Eubacterial 16S | 6.0 (0.3) | 5.9 (0.4) | 6.0 (0.3) | 0.630 |
| Lactobacillus-group | 3.9 (0.9) | 4.2 (0.7) | 3.7 (0.9) | 0.060 |
| Ruminococcus torques 91% | 4.6 (0.8) | 4.4 (0.8) | 4.8 (0.7) | 0.461 |
| Ruminococcus torques 94% | 3.8 (1.0) | 4.0 (1.3) | 3.6 (0.8) | 0.409 |
| Streptococcus bovis-like | 2.7 (1.5) | 3.2 (2.1) | 2.5 (1.2) | 0.732 |
| Veillonella spp. | 3.4 (1.0) | 3.5 (1.1) | 3.4 (1.1) | 0.745 |
| qPCR assay | All (n = 44) | BMI < 251 (n = 17) | BMI > 25 (n = 22) | P2 |
| Bifidobacterium catenulatum/Pseudocatenulatum | 12 | 3 | 7 | 0.315 |
| Bifidobacterium spp. | 44 | 17 | 22 | ND |
| Clostridium coccoides/Eubacterium rectale-group | 44 | 17 | 22 | ND |
| Clostridium cocleatum 88% | 33 | 13 | 15 | 0.568 |
| Collinsella aerofaciens-like | 29 | 15 | 12 | 0.024 |
| Coprococcus eutactus 97% | 16 | 7 | 8 | 0.759 |
| Desulfovibrio desulfuricans-group | 29 | 11 | 15 | 0.819 |
| Eubacterial 16S | 44 | 17 | 22 | ND |
| Lactobacillus-group | 44 | 17 | 22 | ND |
| Ruminococcus torques 91% | 38 | 15 | 18 | 0.582 |
| Ruminococcus torques 94% | 29 | 9 | 16 | 0.202 |
| Streptococcus bovi-like | 33 | 11 | 18 | 0.225 |
| Veillonella spp. | 41 | 16 | 20 | 0.709 |
In linear modeling of continuous data, R. torques 94% was associated with an increase in self-reported bowel symptoms [analysis of variance (ANOVA), P < 0.05]. When the IBS subjects were grouped according to whether R. torques 94% was detected in their fecal samples (Table 7), self-reported bowel symptoms tended to be higher among subjects with R. torques 94% present (ANOVA, P = 0.056). Interestingly, presence of R. torques 94% had a negative effect on the abundance of C. eutactus 97% (P < 0.01), C. aerofaciens-like (P < 0.05) and C. cocleatum 88% (P < 0.05) phylotypes.
| Variable | All samples | Ruminococcus torques 94% | |
| Detected | Undetected | ||
| Symptom sums1 | |||
| All symptoms | 83.6 (20.0) | 87.6 (19.6) | 75.9 (19.1) |
| Bowel symptoms | 24.1 (6.5) | 25.4 (6.5) | 21.5 (5.8)23 |
| Systemic symptoms | 16.4 (4.8) | 17.0 (4.7) | 15.2 (4.8) |
| Social function | 9.3 (4.3) | 10.0 (4.5) | 7.8 (3.6) |
| Emotional function | 33.9 (8.1) | 35.2 (8.0) | 31.5 (7.9) |
| qPCR assays4 | |||
| Bifidobacterium catenulatum/Pseudocatenulatum | 2.5 (1.7) | 2.5 (1.7) | 2.5 (1.6) |
| Bifidobacterium spp. | 5.8 (1.0) | 5.8 (1.0) | 5.9 (1.1) |
| Clostridium coccoides /Eubacterium rectale-group | 7.3 (0.2) | 7.3 (0.2) | 7.3 (0.2) |
| Clostridium cocleatum 88% | 3.9 (2.8) | 3.1 (3.0) | 5.4 (1.7)5 |
| Collinsella aerofaciens-like | 3.0 (3.8) | 1.6 (4.0) | 5.6 (0.7)5 |
| Coprococcus eutactus 97% | 1.9 (2.8) | 0.9 (2.2) | 3.7 (3.0)6 |
| Desulfovibrio desulfuricans-group | 1.9 (3.0) | 1.4 (3.1) | 2.7 (2.6) |
| Eubacterial 16S | 6.0 (0.3) | 5.9 (0.3) | 6.0 (0.4) |
| Lactobacillus-group | 3.9 (0.9) | 3.8 (0.8) | 4.1 (1.0) |
| Ruminococcus torques 91% | 4.1 (1.5) | 4.1 (1.6) | 3.9 (1.3) |
| Ruminococcus torques 94% | 2.7 (1.8) | 3.8 (1.0) | - |
| Streptococcus bovis-like | 1.9 (1.9) | 1.6 (1.6) | 2.4 (2.3) |
| Veillonella spp. | 3.0 (1.7) | 3.2 (1.7) | 3.0 (1.8) |
No other bacterial associations with symptom scores could be verified. Although in particular the C. aerofaciens-like phylotype had a negative association with R. torques 94%, its relationship with self-reported symptoms remained obscure. The phylotype was, however, associated with lower BMI values (Mann-Whitney test for present-absent data; P < 0.01) and lower blood pressure (Mann-Whitney test for present-absent data for systolic and diastolic blood pressure; P < 0.01 and 0.01, respectively), and the data also suggested a link to lower blood sugar levels (P = 0.06). The C. aerofacien-like phylotype was less frequently observed in subjects with BMI above 25 (Table 6) and none of the six subjects with BMI values over 30 had the C. aerofaciens-like phylotype in their feces. Similary to C. aerofaciens, C. eutactus 97% was also associated with lower blood pressure (P < 0.05 and 0.05 for diastolic and systolic blood pressure, respectively). A positive association was present between the presence of C. aerofaciens and amounts of C. eutactus 97% (P < 0.01), whereas a strong negative effect on R. torques 94% was observed (P < 0.001). In addition, when the IBS subjects were categorized according to their BMI, subjects with a BMI value over 25 had more bifidobacteria than normal-weight subjects, but less lactobacilli in an almost significant manner (Table 5).
Self-reported symptoms and GI microbiota composition of IBS patients were analyzed to investigate putative biomarkers for the disorder. Interesting associations between GI microbiota composition and symptom severity were observed.
To measure IBS patients’ symptoms, we applied the IBDQ, designed for assessing the quality of life of IBD patients[16], with some modifications for IBS patients. According to the Spearman’s correlations calculated for each question and symptom group, the questions generally described best the group in which they were included (data not shown). Generally, the range observed for each question contained the entire available scale, indicating that the patients formed a heterogeneous group regarding the severity of individual symptoms (Table 3). However, between symptom groups, there were high correlations, indicating that although the questions measured severity of specific issues, the groups themselves actually measured the same health issues (Table 4). This is understandable since IBS patients’ physical and mental symptoms reflect their well-being at a given point of observation[23]. Bearing in mind that the questionnaire was intended for IBD patients, the results should be interpreted with caution. For example, reasons underlying weight problems are different in IBS patients than in IBD patients, who may experience problems with either loss or gain of weight, depending on the status of their disease[24]. In our study, weight problems were correlated with a higher BMI and can thus be considered a measure of problems with weight. Division of the patients into two groups according to the BMI values resulted in a significant divergence (P < 0.01) in the systemic symptom scores between these two groups, with the patients having a BMI in excess of 25 experiencing more symptoms than leaner subjects. This observation may suggest that some variables other than severity of IBS might be affected by BMI, as seemed to be the case for lactobacilli and bifidobacteria (Table 5). In addition, as some of the participants were treated for hypertension, any connections between blood pressure and GI microbiota should be observed with extreme caution.
C. aerofaciens-like phylotype had interesting associations with patient characteristics. We have previously observed a reduction in the amount of C. aerofaciens in the fecal samples of IBS patients compared with healthy controls[4]. Recently, Mäkivuokko et al[25] reported that elderly subjects using non-steroidal anti-inflammatory drugs (NSAIDs) had reduced amounts of C. aerofaciens present in their feces relative to healthy young subjects and elderly subjects without NSAIDs. A link between anti-inflammatory drugs and the absence of C. aerofaciens was suggested by the authors. In this present study, we observed a negative correlation between the presence (or amounts) of C. aerofaciens and the BMI value of test subjects. Notably, obese (BMI > 30) subjects were negative for C. aerofaciens, but contradictory to our results, Turnbaugh et al[26] have found C. aerofaciens to be more prominent in obese than lean twins and their mothers. Obesity has been associated with a low-grade systemic inflammation in which the GI microbiota may be involved[27]; this could explain the negative association observed for C. aerofaciens and BMI values. It was difficult to conclude whether C. aerofaciens could have any role in IBS; in general, overweight (BMI > 25) subjects reported more systemic symptoms than normal-weight subjects. This leads to a problem in interpretation of the results; as the measured symptom groups correlate strongly with each other, it is difficult to determine whether a rise observed in one symptom group is actually caused by a rise in another symptom group rather than by IBS itself.
The phylogenetically most similar species to R. torques 94% which has previously been associated with IBS-D[18] is a known mucin degrader[28], and the reported increase of mucin in the context of IBS could explain the observed link between this phylotype and the symptoms. The negative association of R. torques 94% with C. cocleatum 88%, C. aerofaciens-like and C. eutactus 97%, observed for this phylotype could thus also be due to sample characteristics (i.e. abundance of mucus and human cells in the samples) and is in correlation with previous results, as these phylotypes have been associated with healthy controls’ GI microbiota in comparison with that of IBS subjects[4]. However, with our knowledge being restricted to the 16S ribosomal DNA sequence of the phylotype, all suggestions about the functions of these bacteria should be considered merely speculative.
Regarding R. torques 94%, an association with BMI values was lacking, while a role for this phylotype in IBS was suggested (Figure 1A and B). Of the bacterial genera and phylotypes here negatively associated with symptom scores or bowel symptoms (Figure 1A and B), Lactobacillus spp., Bifidobacterium spp., D. desulfuricans-group, C. aerofaciens-like and C. eutactus 97% have previously been detected in lowest quantities among IBS-D patients in comparison to other IBS symptom subtypes and healthy control subjects[5,18]. Veillonella spp. has previously been associated with constipation-predominant IBS subjects[5] but was not found to correlate with self reported IBS symptoms in this study.
The observed higher abundance of Bifidobacterium spp. in overweight subjects has previously been reported in a large metagenomic study[26]. Interestingly, an energy-restricted diet has been shown to reduce C. coccoides-group, Bifidobacterium longum and Bifidobacterium adolescentis counts and increase Bacteroides fragilis- and Lactobacillus-group counts in originally overweight adolescents, with the effect being more pronounced among subjects who had lost more weight[29]. However, as Santacruz et al[29] concluded, it may well be the proportional amounts of various bacterial groups within the GI tract rather than their absolute numbers that play a role or react in complex events within the GI tract; they found the Bifidobacterium to C. coccoides-group ratio to increase in correlation with weight loss. In our study, the C. coccoides/E. rectale-group levels were the same in normal-weight and overweight subjects (Table 5).
In conclusion, our findings indicate that certain bacterial phylotypes might serve as markers of symptom severity in IBS. While the presence of R. torques 94% was associated with an increase in symptom severity, some other phylotypes seemed to act in the opposite direction. These microbes are, however, not found in all individuals and they may also be present in healthy subjects’ samples[4]; therefore it is unlikely that their presence or absence in the GI tract would be the underlying cause of IBS.
Irritable bowel syndrome (IBS) is a common functional gastrointestinal (GI) disorder and results in a reduced quality of life. Alterations in the human GI microbiota have been detected among patients suffering from the syndrome. The abnormalities in the GI microbiota are suggested to contribute to IBS symptoms.
The role of GI microbiota in IBS has been under investigation and studies suggest that microbes associated with IBS possess potential as non-invasive biomarkers. Since the majority of the GI bacteria are uncultivable, molecular methods are crucial in this field and have enabled a deeper study of the disturbed microbiota. The authors examined whether the quantities, or presence or absence, of certain microbes previously linked to either IBS or healthy microbiota, correlated with the symptoms experienced by IBS patients.
Alterations in the overall microbiota and certain microbial phylotypes have been detected in IBS. The results of this study suggest that there is a connection between IBS-related microbiota and severity of self-reported symptoms.
The findings in this study indicate that certain bacterial phylotypes are associated with symptom severity in IBS. These bacteria may serve as biomarkers of the course of the condition.
Human intestinal microbiota is the ensemble of all microbes in the gastrointestinal tract. The term bacterial phylotype stands for an operative taxonomic unit determined by the 16S rRNA gene sequence similarity. Quantitative real-time polymerase chain reaction (qPCR) is a method that enables quantification of target DNA molecules in a sample. In this study 16S rDNA sequences of known bacterial genera and of bacterial phylotypes were quantified using qPCR.
This paper represents a large amount of work. Although the results are largely negative, they should be published, since IBS is such an important issue.
Peer reviewers: Tauseef Ali, MD, Assistant Professor, Section of Digestive Diseases and Nutrition, University of Oklahoma Health Sciences Center, 920 SL Young Blvd, Oklahoma City, OK 73104, United States; Dr. William R Parker, PhD, Assistant Professor, Department of Surgery, Duke University Medical Center, Box 2605, Durham, NC 27710, United States
S- Editor Tian L L- Editor Logan S E- Editor Ma WH
| 1. | Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480-1491. [Cited in This Article: ] |
| 2. | Hillilä MT, Siivola MT, Färkkilä MA. Comorbidity and use of health-care services among irritable bowel syndrome sufferers. Scand J Gastroenterol. 2007;42:799-806. [Cited in This Article: ] |
| 3. | Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185-194. [Cited in This Article: ] |
| 4. | Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24-33. [Cited in This Article: ] |
| 5. | Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373-382. [Cited in This Article: ] |
| 6. | DuPont AW. Postinfectious irritable bowel syndrome. Clin Infect Dis. 2008;46:594-599. [Cited in This Article: ] |
| 7. | Chadwick VS, Chen W, Shu D, Paulus B, Bethwaite P, Tie A, Wilson I. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778-1783. [Cited in This Article: ] |
| 8. | Kajander K, Hatakka K, Poussa T, Färkkilä M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther. 2005;22:387-394. [Cited in This Article: ] |
| 9. | Kajander K, Myllyluoma E, Rajilić-Stojanović M, Kyrönpalo S, Rasmussen M, Järvenpää S, Zoetendal EG, de Vos WM, Vapaatalo H, Korpela R. Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther. 2008;27:48-57. [Cited in This Article: ] |
| 10. | Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O’Mahony L, Kiely B, Shanahan F, Quigley EM. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581-1590. [Cited in This Article: ] |
| 11. | Bengtson MB, Rønning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: genes and environment. Gut. 2006;55:1754-1759. [Cited in This Article: ] |
| 12. | Aerssens J, Camilleri M, Talloen W, Thielemans L, Göhlmann HW, Van Den Wyngaert I, Thielemans T, De Hoogt R, Andrews CN, Bharucha AE. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2008;6:194-205. [Cited in This Article: ] |
| 13. | Bian ZX, Li Z, Huang ZX, Zhang M, Chen HL, Xu HX, Sung JJ. Unbalanced expression of protease-activated receptors-1 and -2 in the colon of diarrhea-predominant irritable bowel syndrome patients. J Gastroenterol. 2009;44:666-674. [Cited in This Article: ] |
| 14. | Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657-1664. [Cited in This Article: ] |
| 15. | Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Müller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45 Suppl 2:II43-II47. [Cited in This Article: ] |
| 16. | Irvine EJ, Feagan B, Rochon J, Archambault A, Fedorak RN, Groll A, Kinnear D, Saibil F, McDonald JW. Quality of life: a valid and reliable measure of therapeutic efficacy in the treatment of inflammatory bowel disease. Canadian Crohn’s Relapse Prevention Trial Study Group. Gastroenterology. 1994;106:287-296. [Cited in This Article: ] |
| 17. | Apajalahti JH, Särkilahti LK, Mäki BR, Heikkinen JP, Nurminen PH, Holben WE. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol. 1998;64:4084-4088. [Cited in This Article: ] |
| 18. | Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, Mättö J, Mäkelä L, Palva A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936-5945. [Cited in This Article: ] |
| 19. | Rowan FE, Docherty NG, Coffey JC, O’Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg. 2009;96:151-158. [Cited in This Article: ] |
| 20. | R Development Core Team. R: A Language and environment for statistical computing. Vienna: R Foundation for Statistical Computing 2008; . [Cited in This Article: ] |
| 21. | Fox J, Ash M, Boye T, Calza A, Chang A, Grosjean P, Heiberger R, Kerns GJ, Lancelot R, Lesnoff M, Messad S, Maechler M, Murdoch D, Neuwirth E, Putler D, Ripley B, Ristic M, Wolf P. Rcmdr: R Commander. 2009;. [Cited in This Article: ] |
| 22. | WHO. Obesity: Preventing and Managing the Global Epidemic. Report of a WHO Consultation on Obesity 1998. World Health Organisation, Geneva. [Cited in This Article: ] |
| 23. | Rey E, García-Alonso MO, Moreno-Ortega M, Alvarez-Sanchez A, Diaz-Rubio M. Determinants of quality of life in irritable bowel syndrome. J Clin Gastroenterol. 2008;42:1003-1009. [Cited in This Article: ] |
| 24. | Stjernman H, Grännö C, Bodemar G, Järnerot G, Ockander L, Tysk C, Blomberg B, Almer S, Ström M, Hjortswang H. Evaluation of the Inflammatory Bowel Disease Questionnaire in Swedish patients with Crohn’s disease. Scand J Gastroenterol. 2006;41:934-943. [Cited in This Article: ] |
| 25. | Mäkivuokko H, Tiihonen K, Tynkkynen S, Paulin L, Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br J Nutr. 2010;103:227-234. [Cited in This Article: ] |
| 26. | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-484. [Cited in This Article: ] |
| 27. | DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, Rittmann BE. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460-469. [Cited in This Article: ] |
| 28. | Leitch EC, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007;9:667-679. [Cited in This Article: ] |
| 29. | Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-Matillas M, Campoy C, Moreno LA, Veiga O, Redondo-Figuero C, Garagorri JM. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (Silver Spring). 2009;17:1906-1915. [Cited in This Article: ] |
| 30. | Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166-1177. [Cited in This Article: ] |
| 31. | Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257-266. [Cited in This Article: ] |
| 32. | Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:2578-2585. [Cited in This Article: ] |
| 33. | Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002;68:114-123. [Cited in This Article: ] |