Article Text
Abstract
BACKGROUND/AIMS Alterations in synthesis and breakdown of extracellular matrix components are known to play a crucial role in tissue remodelling during inflammation and wound healing. Degradation of collagens is highly regulated by a cascade of matrix metalloproteinases (MMPs). The current study was therefore designed to determine gene expression patterns of MMPs and their tissue inhibitors (TIMPs) in single endoscopic biopsies of patients with inflammatory bowel disease (IBD).
PATIENTS/METHODS mRNA expression was measured by quantitative competitive polymerase chain reaction (PCR) in biopsies from patients with ulcerative colitis (n=21) and Crohn's disease (n=21). Protein expression was analysed by western blotting and immunohistochemistry.
RESULTS MMP-2, MMP-14, and TIMP-1 mRNAs were marginally increased in inflamed, but 9–12-fold increased in ulcerated colonic mucosa in IBD whereas TIMP-2 mRNA expression remained unchanged. MMP-1 and MMP-3 mRNA expression correlated well with the histological degree of acute inflammation, resulting in more than 15-fold increased MMP-1 and MMP-3 mRNA levels in inflamed versus normal colon samples from patients with ulcerative colitis and Crohn's disease.
CONCLUSION Profound overexpression of MMP-1 and MMP-3 mRNA transcripts suggests an important role for these enzymes in the process of tissue remodelling and destruction in inflammatory bowel disease.
- matrix metalloproteinases
- polymerase chain reaction
- extracellular matrix
- Crohn's disease
- ulcerative colitis.
Abbreviations used in this paper
- MMP
- matrix metalloproteinase
- MT-MMP
- membrane-type MMP
- TIMP
- tissue inhibitor of matrix metalloproteinases
- PCR
- polymerase chain reaction
- IBD
- inflammatory bowel disease
- ECM
- extracellular matrix
- CD
- Crohn's disease
- UC
- ulcerative colitis
- mRNA
- messenger RNA
- cDNA
- complementary DNA
- APAAP
- alkaline phosphatase/anti-alkaline phosphatase
- RT
- reverse transcription
- PVDF
- polyvinyldifluoride
- ECL
- enhanced chemiluminescence
Statistics from Altmetric.com
- matrix metalloproteinases
- polymerase chain reaction
- extracellular matrix
- Crohn's disease
- ulcerative colitis.
Chronic mucosal inflammation is characterised by an inflammatory cell infiltrate associated with changes in epithelial proliferation and migration,1 paralleled by intensive remodelling of the subepithelial connective tissue leading to increased turnover of extracellular matrix (ECM) components.2Coordinate synthesis and degradation of ECM components is necessary for many important pathophysiological processes involved in inflammation, such as leucocyte invasion,3 epithelial migration,4 ,5 neovascularisation,6 and wound healing.7 Disturbance of the balance between synthesis and degradation of ECM components may result either in progressive organ destruction, as seen for example in ulcer formation,8 or excessive deposition of collagens resulting in fibrosis.2 ,9
Degradation of ECM components is tightly controlled by the enzymatic activity of matrix metalloproteinases (MMP). At least 17 different MMPs have been identified in humans (MMP 1–20, numbers 4–6 were not used) (for recent reviews see Brown10 and Massova and colleagues11). Depending on structure and substrate specificity, MMPs are further subdivided.12 MMP-1, MMP-8, and MMP-13, termed collagenases, are capable of cleaving triple helical interstitial collagen types I, II, III, and X and play a key role in degradation of the interstitial ECM.13 The gelatinases, MMP-2 and MMP-9, digest denatured collagens as well as collagen type IV and are implicated in cell invasion.14 Stromelysins (MMP-3 and MMP-10) have a broad spectrum of substrates such as proteoglycans, collagens, gelatins, laminin, fibronectin, and elastin. Moreover, these enzymes can activate collagenases by proteolytic cleavage of the pro-domain. The most recently discovered group comprise four membrane bound matrix metalloproteinases (MMP-14 to MMP-17) called membrane-type MMPs (MT-MMP) with a transmembrane domain at their carboxy terminus. In contrast with soluble MMPs, these enzymes are processed intracellularly and expressed in their active form. Substrate specificity for MT-MMP-1 (MMP-14) has been described for interstitial collagens types I, II, and III, fibronectin, laminin, and proteoglycans.15 Even more important is that MMP-14 can bind and activate MMP-2 and MMP-13 (for reviews see Sato and Seiki16 and Nagase17).
As one might expect for these potentially harmful enzymes, their enzymatic activity is tightly controlled. Soluble MMPs (with the exception of MMP-11) are secreted as latent pro-MMPs and are activated either by stepwise proteolytic removal of the amino terminal domain or by treatment with SH reactive or denaturing agents.17After initial cleavage by trypsin or plasmin, MMP-1 can be activated by active MMP-3 and, to a lesser extent, by MMP-2, MMP-10, and MMP-7. Furthermore, MMP-14 has been shown to be an important activator of MMP-2.18 Increased membrane expression of MT-MMPs initiates a cascade of MMP activation and results in increased pericellular fibrinolysis, an important feature in cell invasion.16
MMP activity is counterbalanced by endogenous inhibitors such as α2 macroglobulin and specific tissue inhibitors of matrix metalloproteinases (TIMP). Currently, four different TIMPs have been identified, revealing different tissue and cell type specific expression and regulation patterns (for review see Gomez and colleagues19). TIMPs inhibit active MMPs by 1:1 stoichiometric binding and the ratio of active MMPs to TIMPs controls collagenolytic activity.
Crohn's disease (CD) and ulcerative colitis (UC) are chronic inflammatory bowel diseases of unknown aetiology. Features of CD include transmural inflammation, fibrosis, and fistula formation, whereas inflammation in UC is limited to the mucosal layers of the colon and rectum. Development of fibrotic strictures leading to intestinal obstruction is a frequent complication in CD,20but this complication is rarely seen in UC.21 The pathophysiological mechanisms underlying fibrotic strictures in CD are currently poorly understood.
To gain further insight into the molecular mechanisms which might be involved in the tissue remodelling process in inflammatory bowel disease (IBD), we analysed expression of matrix degrading MMPs and their inhibitors. Reliable measurement of the in situ enzymatic activity of MMPs is limited for technical reasons as any extraction procedure might either be incomplete or destroy the binding of MMPs to their inhibitors.22 Furthermore, MMPs are easily degraded. Several studies have shown that expression of MMPs is mainly transcriptionally regulated and that MMP protein concentrations correlate well with mRNA expression in vitro.23-25 We therefore established a competitive quantitative polymerase chain reaction (PCR) to simultaneously quantitate mRNA expression of MMP-1 and MMP-2, as well as their major activators MMP-3 and MMP-14, and of tissue type inhibitors TIMP-1 and TIMP-2 in single endoscopic biopsies.
Methods
MATERIALS
The following reagents were purchased: ECL western blotting analysis system from Amersham, Buckinghamshire, UK; Bradford protein assay from Bio-Rad Lab., Hercules, California, USA; agarose from Biozym, hess. Oldendorf, FRG; DNase I, oligo-dT-primer, and the SuperScript Preamplifikation-Kit from Gibco, Rockville, USA; dNTPs and Taq-polymerase from Pharmacia, Uppsala, Sweden; RNeasy RNA extraction Kit, QIAshredder Kit, and Qiaquick gel extraction Kit from Qiagen, Santa Clarita, USA.
Antibodies: α-MMP1 polyclonal rabbit IgG was purchased from Biogenesis, Pool, UK; α-MMP-1 clone 36665.111 monoclonal mouse IgG1 and α-MMP-3 (clone 10D6) monoclonal mouse IgG1 from R&D systems, Wiesbaden, FRG; α-MMP-2 (clone Ab-3) monoclonal mouse IgG1 from Oncogen Research Products, Cambridge, USA; α-MMP-14 (clone RP 63) polyclonal rabbit IgG (kind gift of K M Miller at British Biotech, Oxford, UK); anti-β-actin (clone ac-15) monoclonal mouse IgG1 from Sigma-Aldrich, Deisenhofen, FRG; and anti-CD68 antibody PGM-1 from Dakopatts, Denmark.
TISSUE SAMPLES
After obtaining informed consent, biopsies were taken at routine diagnostic colonoscopies from patients with IBD. Biopsies were taken from areas which macroscopically demonstrated the highest degree of inflammation in the closest possible proximity to those biopsies taken for routine histopathological examinations. Biopsies were immediately snap frozen in liquid nitrogen after removal. If there were areas of normal appearing colonic mucosa, these were also biopsied. Biopsies from inflamed colonic mucosa of 45 different patients with IBD (22 patients with CD and 23 patients with UC) were analysed. A control group of 14 biopsies from these patients (seven patients with CD and seven with UC) appeared macroscopically normal and showed no signs of inflammation at histopathological examination. This study was approved by the local ethics committee.
RNA PURIFICATION AND cDNA CONSTRUCTION
Total RNA was isolated from single biopsies using the RNeasy Kit (Qiagen, Santa Clarita, USA) following the manufacturer's instructions. In brief, tissue samples were mechanically disrupted under liquid nitrogen and freed from ice. The frozen tissue (8–10 mg) was lysed in 350 μl of lysis buffer containing guanidinium salt as an RNAse inhibitor. Tissues were lysed for 3×10 seconds using an ultrasonic disintegrator (Sonoplus HD 70, Bandelin Electronic, Berlin, FRG) and homogenised using the QIAshredder Kit (Qiagen, Santa Clarita, USA). After centrifugation, total RNA was purified from the supernatant by affinity chromatography using the RNeasy columns. Aliquots of 1.5 μg of total RNA were further purified by DNAse I (Gibco, Rockville, USA) digestion and reverse transcribed into cDNA using the oligo-dT-primers and the SuperScript Preamplifikation-Kit (Gibco, Rockville, USA).
POLYMERASE CHAIN REACTION (PCR)
PCR reactions were carried out in a total volume of 50 μl containing primer (400 nM each), dNTPs (Pharmacia, Uppsala, Sweden; 200 μM of each dNTP), KCl (50 mM), MgCl2 (1.5 mM), Tris (10 mM), and 1 unit of Taq polymerase (Pharmacia, Uppsala, Sweden). PCR was performed in a Peltier thermocycler (PTC-200, MJ-Research, USA) under the following conditions: initial heating to 95°C for five minutes, then 30 cycles (35 cycles for MMP-1 or MMP-2) at 94°C for 40 seconds, 63°C for 60 seconds and 72°C for 60 seconds, followed by 72°C for a further seven minutes.
Primer sequences were chosen using the commercial PcGene software package (Intelligenetics, Geneva, Switzerland) on published cDNA sequences retrieved via the Internet from the GeneBank database. Selected primer sequences are given in table 1.
Primer sequences used
REVERSE TRANSCRIPTION COMPETITIVE PCR
The principle of a reverse transcription (RT) competitive PCR is coamplification of a known amount of an artificial DNA (so-called competitor) with different amounts of cDNA. If the amplification efficiencies of this competitor and the original PCR template are equal, it is possible to determine the cDNA concentration of a specific PCR template by measuring the ratio of competitor and wild-type PCR products.
Competitors were constructed using a “semi-nested” PCR technique as described by Jin and colleagues.26 The wild-type PCR products were reamplified using combined (so-called mimic) sense primers, which bind 80–120 bp downstream of the original sense primer but contain the original sense primer sequence at their 5′ ends. This results in 10–20% shorter amplificates than the wild-type PCR products that contain the same primer binding sequences at their ends. These competitor constructs were purified by agarose gel electrophoresis followed by gel extraction using the commercial Qiaquick gel extraction kit (Qiagen, Santa Clarita, USA).
For quantitative PCR, five serial 1:3 dilutions of cDNA derived from one reverse transcription were coamplified with fixed amounts of one of the different competitors. After amplification, the wild-type and competitor PCR products were separated in 2% agarose (Biozym, hess. Oldendorf, FRG) gels and stained with ethidium bromide. Digital images of the stained gel were taken using the QuickStore-plus-II system (MS Laborgeräte, Heidelberg, FRG) with an integrated UV light tray and a CCD camera, and densitometry of PCR products was performed using the ScionImage software package (Scion Corporation, Frederick, Maryland, USA.). The logarithm of the density ratios (wild-type/competitor) was plotted against the logarithm of cDNA dilution. At the competition equivalence point (log (density ratio)=0), the initial target cDNA is diluted to the concentration (w/v) of competitor added. This allows calculation of the molar concentration of the target wild-type PCR product in the cDNA. Expression of the different target sequences studied was normalised to expression of β-actin message in the same cDNA sample.
WESTERN BLOTS
Single colonic biopsies (typically 10 mg of tissue) were lysed in a lysis buffer (150 mM NaCl, 2 mM EDTA, 20 mM Tris HCl (pH 7.8)), 50 mM β-glycerolphosphate, 0.5% TNP 40, and 1% glycerin containing a cocktail of the following protease inhibitors: 1 mM sodium orthovanadate, 1 mM dithiothreitol, 10 kIU/ml aprotinin, 10 mM sodium fluoride, 2 μM leupeptin, and 2 mM phenylmethylsulphonamide. Tissue was lysed on ice using an ultrasonic homogeniser (Sonoplus HD 70, Bandelin Electronic, Berlin, FRG). After centrifugation, protein concentration was measured in the supernatant using the colorimetric Bradford protein assay (Bio-Rad Lab., Hercules, California, USA). Total protein (20 μg) was separated by SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) using 12% polyacrylamide gels. The separated proteins were electroblotted onto polyvinyldifluoride (PVDF) membranes. Non-specific binding sites were blocked by incubating the membranes for one hour at room temperature in a solution of 5% (w/v) non-fat dry milk in phosphate buffered saline containing 0.1% of the detergent Tween 20 (PBS-T). After washing, membranes were incubated with monoclonal antibodies specific for MMP-1 (5 μg/ml), MMP-2 (1 μg/ml), or MMP-3 (10 μg/ml) at 4°C overnight.
After washing, bound antibodies were detected by enhanced chemiluminescence (ECL) using the commercial ECL western blotting analysis system (Amersham, Buckinghamshire, UK) according to the manufacturer's instructions. To confirm equivalent protein loading, membranes were stripped by incubation in 62.5 mM of Tris HCl (pH 6.8), 2% SDS, and 100 mM 2-mercaptoethanol at 55°C, subsequently washed with PBS-T, and reprobed with anti-β-actin antibody (dilution 1: 2500).
IMMUNOHISTOCHEMISTRY
Immunohistochemical analysis of formalin fixed and paraffin embedded tissue biopsies taken at the same colonic area as the biopsies analysed by competitive RT-PCR was carried out using the alkaline phosphatase/anti-alkaline phosphatase (APAAP) method.27After pressure cooker antigen retrieval, sections were incubated at room temperature for 30 minutes with a rabbit polyclonal antibody against MMP-1 (Biogenesis, Pool, UK) using a 1:5 dilution. Sections were subsequently treated with a mouse antirabbit antibody (M737 Dakopatts, Denmark) and then with a rabbit antimouse antibody (Z259 Dakopatts, Denmark) for 30 minutes each. Finally, slides were incubated with APAAP complex 1:100 (Dakopatts, Denmark). New fuchsin was used as the developer. The sections were counterstained with haematoxylin and mounted using Kaiser's gelatin solution (Merck, Germany). To identify the cells which stained positive for MMP-1, double immunostaining using the avidin-biotin-peroxidase method28 before APAAP staining was performed. After treatment with 0.3% hydrogen peroxide to remove endogenous peroxidase, sections were first incubated for 30 minutes with anti-CD68 antibody (PGM-1, Dakopatts, Denmark) diluted 1:100. Sections were then incubated using a biotinylated sheep antimouse antibody and the avidin-peroxidase complex (both from the commercial KIT 5001, Dakopatts, Denmark). Peroxidase staining was developed using 3,3 diaminobenzidine tetrahydrochloride and a hydrogen peroxide solution (Sigma-Aldrich, Deisenhofen, FRG). After digestion with pronase 0.1% for 10 minutes, APAAP staining for MMP-1 was performed as described above.
HISTOLOGICAL GRADING
The degree of acute inflammation was graded as suggested by Truelove and Richards29 on a four point scale: 0 (normal), no significant inflammation; 1 (mild), elevated number of mucosal leucocytes but intact epithelium; 2 (moderate), aggregates of leucocytes with crypt abscesses and erosions but no ulcerations of the epithelium; and 3 (severe), significant ulceration of the epithelium by mononuclear cell infiltrate. Histological grading, performed by an experienced pathologist (SEC) without knowledge of the endoscopic reports or experimental data, was conducted on sections of biopsies taken at the closest possible proximity to the biopsies analysed by RT competitive PCR.
STATISTICAL ANALYSIS
Statistical analysis was performed using the PRISM Statistic Package, version 2.0 (GraphPad Software Inc. San Diego, California, USA). Comparisons between three or more groups were performed using the Kruskal-Wallis test (non-parametric analysis of variances). The groups were considered different at a p value <0.05. In these cases p values for comparisons between groups were calculated using Dunn's multiple comparisons as a non-parametric post-test. Correlations were computed using the Spearman rank test.
Results
ESTABLISHMENT OF A COMPETITIVE PCR FOR MATRIX METALLOPROTEINASES
To study the metabolism of the ECM in single endoscopic biopsies from IBD patients, we developed an RT competitive PCR for the matrix metalloproteinases (MMP-1, -2, -3, and -14) as well as for their tissue inhibitors TIMP-1 and TIMP-2. As a marker for synthesis of the ECM, we studied mRNA expression of the alpha-1 chain of procollagen type III. As the process of RNA purification from biopsies and the following reverse transcription into cDNA cannot be reliably controlled for in this system, expression of different genes was normalised to β-actin and GAPDH transcripts in the same cDNA sample. For validation of the competitive PCR, equal amplification efficiencies of the gene specific PCR products and the PCR competitors had to be shown. Figure 1demonstrates equal amplification efficiency of the final MMP-1 competitor and the corresponding MMP-1 PCR product. The ratio of competitor to PCR product remains constant after PCR amplification has reached a plateau after more than 30 cycles. These validation procedures were performed for each transcript analysed in this study (data not shown).
Evaluation of equal amplification efficiencies of the MMP-1 PCR product and the MMP-1 competitor construct. Purified MMP-1 PCR product (+; 247 bp) and the MMP-1 competitor construct (#; 222 bp) were mixed in an OD ratio of 1:1 (lane 1) and 1:5 (lane 6). These solutions were diluted 1:200 000 and used as templates for PCR for 27 cycles (lanes 2, 7) and 35 cycles (lanes 3, 8). An inverted image of an ethidium bromide stained gel is shown demonstrating equal density ratios before (lanes 1, 6) and after (lanes 2, 3, 7, 8) reamplification. Lanes 4 and 5 contain a DNA mass standard used for calibration of the densitometry. Numbers on the right indicate the DNA size in base pairs (bp).
INCREASED EXPRESSION OF PROCOLLAGEN TYPE III mRNA IN INFLAMED COLONIC MUCOSA OF IBD PATIENTS
Using a competitive PCR technique, median procollagen type III (alpha-1 chain) mRNA expression was 4.3% of the β-actin mRNA transcripts in non-inflamed colonic mucosa of IBD patients. In inflamed colon samples, there was a significant increase in median collagen type III expression to 12.9% of β-actin transcripts in CD and 21.1% of β-actin transcripts in UC (fig 2G).
Expression of MMP-1, -2, -3, -14, the tissue inhibitors TIMP-1 and TIMP-2, collagen type III, and GAPDH mRNA in colon biopsies from non-inflamed areas and inflamed areas of patients with Crohn's disease (CD) and ulcerative colitis (UC). Expression was measured by a competitive PCR technique and is expressed as the molar ratio of gene specific to β-actin copies. Scattergrams of all measurements in a logarithmic scale are shown. Bars represents the medians of the groups. Degree of statistical significance calculated by the Kruskal-Wallis test is given in each upper left corner. Statistically significant differences between the three groups (*p<0.05, **p<0.01, ***p<0.001; NS, not significant) were calculated using Dunn's post hoc test.
INCREASED EXPRESSION OF MATRIX METALLOPROTEINASES IN INFLAMED COLONIC MUCOSA OF IBD PATIENTS
In non-inflamed colonic mucosa, median expression of MMP-2 and MMP-14 mRNA was 0.27% and 0.18% of β-actin expression (fig 2B, D). In contrast, median mRNA expression of MMP-1 and MMP-3 was lower (5×10−6 and 8×10-5 of β-actin transcripts, respectively). MMP-1 could not be detected in six of the 14 non-inflamed tissue samples studied. In inflamed colonic mucosa of IBD patients, we observed a highly significant increase in the expression of MMP-1, -2, -3, and -14 (fig 2A–D). Median expression of MMP-1 increased 20-fold in CD (to 1×10-4) and 42-fold (to 2.2×10-4) in UC compared with non-inflamed samples. The increase in MMP-3 expression was 15-fold (to 116×10-5) in CD and 43-fold (to 334×10-5) in UC. The increase in the expression of MMP-2 and MMP-14 mRNA, although statistically significant, was much less pronounced. The increase in median expression was 1.5-(2.2-) fold in CD and 3.1-(3.6-) fold in UC for MMP-2 and MMP-14, respectively.
This increased expression of MMP-1, MMP-2, and MMP-3 was also noticed at the protein level. Western blotting of whole tissue lysates demonstrated increased expression of MMP-1, MMP-2, and MMP-3 in lysates from inflamed colonic mucosa (fig 3; lanes 2, 4, 6) compared with non-inflamed colonic mucosa from the same patients (fig 3; lanes 1, 3, 5). Most intense bands for all three MMPs were in the latent (inactive) form. With respect to MMP-2, only the latent form of this matrix metalloproteinase was detected, with an approximate molecular weight of 72 kD. For MMP-1 and MMP-3, in addition to the latent MMPs at about 56 kD and 60 kD, respectively, there were also minor forms detectable (at about 45 kD and 48 kD, respectively) which correspond to the known molecular weights of active MMP-1 and MMP-3.
Increased protein expression of MMP-1, MMP-2, and MMP-3 in inflamed (i) versus non-inflamed (n) colon biopsies from patients with IBD. Endoscopic biopsies from macro- and microscopically normal (lanes 1, 3, 5) and inflamed (lanes 2, 4, 6) colon of one patient with Crohn's disease (lanes 1, 2) and two patients with ulcerative colitis (lanes 3–6) were lysed in a buffer containing a cocktail of protease inhibitors. Proteins were separated by polyacrylamide gel electrophoresis and transferred to PVDF membranes. Membranes were probed with antibodies against MMP-1 (A), MMP-2 (B), and MMP-3 (C) and bound antibody was detected by enhanced chemiluminescence. Membranes were reprobed with an antibody against β-actin (D) as a loading control. Estimated molecular weights of the bands detected are given on the right.
INCREASED EXPRESSION OF TIMP-1 BUT NOT TIMP-2 mRNA IN THE INFLAMED COLONIC MUCOSA OF IBD PATIENTS
Competitive PCR revealed median TIMP-1 expression of 2.0% of β-actin transcripts and TIMP-2 expression of 0.44%. In the inflamed colon of IBD patients, there was a moderate but significant increase in median TIMP-1 mRNA expression to 4.2% in CD (2.1-fold increase) and 7.9% in UC (3.9-fold increase). In contrast, as shown in figure 2F, median expression of TIMP-2 was not significantly altered (median expression 0.56% in CD and 0.73% in UC).
TIMP-1 mRNA expression demonstrated a good correlation with collagen type III expression (Spearman r=0.61) as well as MMP-14 (r=0.55) and MMP-2 (r=0.5) whereas the correlation with MMP-3 and TIMP-2 (Spearman r=0.43 and 0.41, respectively) was weak; there was no correlation with MMP-1 expression.
As TIMP-1 is known to inhibit the enzymatic activity of the MMPs studied, we also calculated MMP/TIMP-1 mRNA ratios. As shown in fig 4B and 4D, the increase in the expression of TIMP-1 mRNA paralleled increased expression of MMP-2 and MMP-14 resulting in equal mRNA ratios of MMP-2/TIMP-1 and MMP-14/TIMP-1 in inflamed and non-inflamed colonic mucosa from IBD patients. In contrast, the increase in expression of MMP-1 and MMP-3 exceeded the increase in TIMP-1 mRNA expression in the inflamed colon. In comparison with non-inflamed colon, the median ratio of MMP-1/TIMP-1 mRNA was 8.8-fold higher in CD and 22.8-fold higher in inflamed UC. The median mRNA ratio of MMP-3/TIMP-1 was increased 10-fold in CD and 13-fold in UC (fig 4A, C).
Molar ratios of matrix metalloproteinase to TIMP-1 specific cDNA derived from reverse transcribed mRNA isolated from colon biopsies of non-inflamed areas and inflamed areas of patients with Crohn's disease (CD) and ulcerative colitis (UC). Competitive PCR measurements are displayed as scattergrams in a logarithmic scale. Bars represent the medians of the groups. Statistical significance, calculated using the Kruskal-Wallis test, is given in each upper left corner. Statistically significant differences between the three groups (*p<0.05, **p<0.01, ***p<0.001; NS, not significant) were calculated using Dunn's post hoc test.
CORRELATION OF MMP AND TIMP mRNA EXPRESSION WITH THE HISTOLOGICAL DEGREE OF ACUTE INFLAMMATION
The degree of acute inflammation was graded according to Truelove and Richards29 on a four point scale: not inflamed (0), mild (1), moderate (2), and severe (3) inflammation, as detailed in the methods section. Histological grading was performed on sections of biopsies taken in closest possible proximity to the biopsies analysed by RT competitive PCR. There was a good correlation between the histological degree of acute inflammation and mRNA expression of collagen type III and MMP-1, MMP-2, and MMP-3 measured by competitive PCR (r>0.5 using the Spearman rank correlation; p<0.0001) (fig 5). For MMP-14 and TIMP-1, the correlation, although significant, was weak (r=0.42). When we compared MMP expression between various degrees of inflammation, the strongest expression of MMPs as well as collagen type III was noted in severely inflamed tissues characterised by ulceration of the epithelium (histological score 3).
Relationship between histological degree of acute inflammation in colon biopsies from IBD patients and mRNA expression of MMPs, TIMP-1, and collagen type III measured by competitive PCR. Scattergrams show the ratio of gene specific transcripts to β-actin transcripts in a logarithmic scale and bars mark the medians of each group. The histological degree of acute inflammation was graded according to Truelove and Richards on a four point scale: no (0), mild (1), moderate (2), and severe (3) inflammation, as outlined in the methods section. Correlation between histological score and mRNA expression was computed using the Spearman rank test and the correlation coefficient r is given in the upper left corner. Statistical significant differences between probes with and without histological confirmed acute inflammation were calculated (*p<0.05, **p<0.01,***p<0.001).
For MMP-2, MMP-14, and TIMP-1, median mRNA expression in these severely ulcerated tissue samples was increased 9–12-fold compared with non-inflamed colonic mucosa. In inflamed tissue samples without ulcerations (histological score 2) this increase was much weaker (2.2–2.7-fold) and was no longer statistically significant.
In contrast, median expression of MMP-1 and MMP-3 was elevated by 230-fold (MMP-1) and 150-fold (MMP-3) in the presence of ulcerations compared with normal mucosa. Also, in tissue samples showing a dense infiltrate of inflammatory cells but no major ulcerations (histological score 2) this increase was pronounced (22–36-fold for MMP-1 and MMP-3, respectively) and was statistically significant (fig 5A, 5C).
No correlation was noted between histological score and expression of TIMP-2 or GAPDH.
COMPARISON OF MATRIX METABOLISM IN CROHN'S DISEASE AND ULCERATIVE COLITIS
As shown in fig 2, mRNA expression of MMPs and collagen type III tended to be higher in inflamed colonic mucosa from UC patients compared with CD which reached statistical significance for collagen type III, MMP-2, and MMP-3. As discussed above, the highest expression of most mRNAs was seen in severely ulcerated tissue samples. As five of the six ulcerated tissue samples originated from patients with UC and only one from a patient with CD, these ulcerated tissue samples were excluded from the analysis to compare tissue samples with a similar histological degree of inflammation. In doing so, median mRNA expression of MMP-1 (CD: 1×10−4/ UC: 1×10−4 of β-actin transcripts) and collagen type III (CD: 12.9%/UC: 12.6% of β-actin transcripts) was equivalent in inflamed colonic mucosa from both entities. Median expression of MMP-2, MMP-3, MMP-14, and TIMP-1 was still 1.5–3.5-fold higher in inflamed tissue samples from patients with UC compared with CD, but this was no longer statistically significant.
IMMUNOHISTOCHEMICAL LOCALISATION OF MMP-1 AND MMP-3 PROTEIN
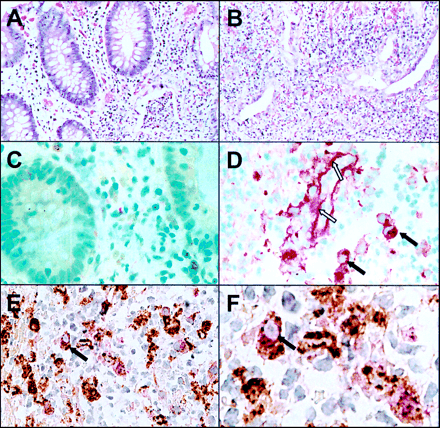
As PCR analysis revealed the strongest over- expression for MMP-1 and MMP-3 in IBD, we investigated the cellular distribution of these enzymes. Using the APAAP and PAP technique on sections of paraffin embedded colon biopsies, positive staining for MMP-1 or MMP-3 was observed only in a few mononuclear cells within the mucosa of the normal colon (see fig 6C). In inflamed colon biopsies of IBD, the number of these positive staining cells was slightly increased but the distribution within the mucosa was patchy. In cases of severe acute inflammation with ulceration there were aggregates of cells staining positive for MMP-1. MMP-1 staining was also detected along the basal membrane of small blood vessels within the inflamed tissue (see fig6D) whereas no MMP-1 staining of blood vessels was observed in non-inflamed tissues.
Immunohistochemical staining of MMP-1 in inflamed and non-inflamed colon of a patient with ulcerative colitis. Section of paraffin embedded biopsies from non-inflamed (A, C) and inflamed (B, D-F) colon were stained with haematoxylin/eosin (A, B) and anti-MMP-1 polyclonal antibody using the APAAP technique (C, D). Double immunostaining with anti-MMP-1 (stained red) and anti-CD68 (stained brown) is shown in (E) and a magnified view is given in (F). Dark arrows indicate macrophages expressing CD68 and MMP-1; open arrows indicate extracellular MMP-1 staining along the basal membrane of a blood vessel.
To identify the mononuclear cells which stained positive for MMP-1, we performed double immunostainings using an anti-CD 68 antibody, which specifically recognises macrophages. As shown in fig 6E and fig 6F, MMP-1 expression (stained red with the APAAP technique) was seen only in cells expressing CD68 (stained brown with PAP technique). The mononuclear cells which stained positive for MMP-1 were therefore identified as macrophages. Similar results were obtained for the immunohistochemical analysis of MMP-3 (data not shown).
Discussion
Using a competitive PCR technique we have established a system for rapid and simultaneous measurement of multiple genes involved in the metabolism of the ECM within a single colonic biopsy. To gain further insight into their biological function in inflammatory bowel disease we analysed relative mRNA expression of collagen type III, matrix metalloproteinases MMP-1 and MMP-2, their respective major activators MMP-3 and MMP-14, as well as their physiological inhibitors TIMP-1 and TIMP-2 in endoscopic biopsies from inflamed and non-inflamed colon of IBD patients.
We observed that mRNA expression of all MMPs was significantly increased in inflamed compared with non-inflamed colonic mucosa of these patients. This increase was most pronounced in ulcerated colonic mucosa. By performing westerns blots, increased expression of MMP-1, MMP-2, and MMP-3 was also demonstrated at the protein level, which supports previous in vitro studies showing a close correlation between mRNA and protein expression of MMPs.25 ,30 ,31
Expression of MMP mRNA transcripts demonstrated a close correlation with the histological degree of inflammation and with collagen type III mRNA expression, which is also significantly increased in the inflamed bowel of IBD patients, supporting previous semiquantitative findings by in situ hybridisation.32 The strongest correlation in our study was noted between procollagen type III and MMP-2 mRNA expression (Spearman r=0.79; p<0.0001). A similar correlation between MMP-2 and procollagen type I mRNA expression has previously been noted in granulation tissue of ulcerated gastric cancer.33
We report here for the first time increased mRNA expression of MMP-14, the membrane bound activator of MMP-2, predominantly in ulcerated colonic mucosa of IBD patients. Inflammation without ulceration causes only a minor (2–2.5-fold) increase in both mRNA steady state levels. In ulcerated tissue samples however, both mRNA levels were increased 9–12-fold compared with non-inflamed mucosa. In a rat model of skin wound healing, MMP-14 was highly expressed by MMP-2 producing fibroblasts located in the granulation tissue of healing wounds.34 A similar coordinate expression of MMP-2 and MMP-14, resulting in increased activation of MMP-2, is seen in cholestatic liver injury35 and is associated with increased turnover of the ECM at the tumour/stroma interface in cancer.36 Expression of MMP-14 is also required for neoangiogenesis,6 which is an important factor in ulcer healing.
We also observed significantly increased expression of TIMP-1 mRNA in inflamed and especially ulcerated colon mucosa from IBD patients, whereas TIMP-2 mRNA levels remained unchanged. Expression of TIMP-1 and TIMP-2 by various cell types is regulated differentially by cytokines and other stimuli in vitro37 ,38 which might be partly explained by the observation that analysis of the promoter sequence of TIMP-2 demonstrated features often seen in housekeeping genes.39
In contrast with MMP-2 and MMP-14, mRNA expression of MMP-1 and MMP-3 was low in normal colonic mucosa. Even using highly sensitive PCR, MMP-1 could not be detected in six of 14 colon biopsies without inflammation. Using the quantitative PCR approach, we demonstrated a significant increase in MMP-1 and MMP-3 mRNA in inflamed colon from IBD patients with a significant correlation between the extent of MMP expression and the histological degree of inflammation. In part, this substantiates earlier findings by Bailey and colleagues.40However, this group did not note changes in MMP-1 expression in IBD. In contrast, in situ hybridisation studies demonstrated increased expression of MMP-1 in inflamed41 and ulcerated42 mucosa from IBD patients.
Using paraffin embedded sections, we were unable to detect significant extracellular staining of MMP-1 or MMP-3, despite positivity for MMP-1 along the basal membrane of blood vessels. In agreement with previous studies,40 we observed intracellular staining of MMP-1 and MMP-3 in mononuclear cells infiltrating the lamina propria. Using double immunostaining we identified these cells as macrophages. It is well known that macrophages are a major source of MMP-1 and MMP-33 ,43 and that T cells can induce large amounts of MMP production in macrophages.44 However, in situ hybridisation studies suggest that α-actin positive cells such as myofibroblasts are a main source of MMP mRNAs in the inflamed gut.41 ,45 ,46 One possible explanation for these differences between immunohistochemistry and in situ hybridisation might be that MMPs produced by α-actin positive cells are not stored in these cells, and after secretion diffuse into and bind to the ECM and are therefore rarely detectable by immunohistochemistry. In contrast with most other cell types, macrophages can store MMP protein, which allows immunohistochemical detection.47
One major new finding of this study is that mRNA expression of MMP-1 and MMP-3 is overly increased compared with the moderately increased expression of their inhibitor TIMP-1 in inflamed mucosa of IBD patients. However, when interpreting these results two considerations need to be kept in mind: (i) although PCR is a powerful tool to quantitate individual messenger RNAs, the biological significance of the proposed counterbalancing of MMPs requires confirmation by net matrix degrading activity, ideally performed in vivo, but limited by technical constraints; (ii) the large number of infiltrating inflammatory cells might secrete relevant proteases which are not detected by PCR aimed at quantitation of transcriptional alterations.
There are several lines of evidence to suggest that elevated MMP-1 and MMP-3 expression may reflect acute tissue damage rather than wound healing. Firstly, MMP-1 and MMP-3 have been shown to cause extensive tissue damage in explants of human fetal gut which was prevented by a synthetic inhibitor of MMP-3.48 Secondly, in coeliac disease, expression of MMP-1 and MMP-3 is increased in active disease and decreases in the healing phase under a gluten free diet.46 Thirdly, in gastric ulcers, the amount of MMP-1 expressing cells correlates with the risk of perforation.49 Fourthly, MMP-1 and MMP-3 expression was increased in chronic intestinal ulcers in IBD and chronic cutaneous ulcers as estimated by in situ hybridisation.42 ,50Fifthly, in a rat model of skin wound healing, highest MMP-1 levels were noticed within 24 hours after initiation of the wound and MMP-1 levels decreased during the healing phase whereas MMP-2 and MMP-14 increased.34
In contrast, treatment with plasmin, a known activator of both MMP-1 and MMP-3, also promotes wound healing in the skin,51 and it has been shown that increased expression of MMP-1 enables keratinocytes to migrate over collagen gels and therefore promote wound healing.5 However, the situation in the intestinal tract may be different. In situ hybridisation studies clearly demonstrate that MMP-1 and MMP-3 are expressed by myofibroblasts and macrophages at the ulcer bed but not by epithelial cells in the inflamed colon.41 ,42 ,46 In addition, de novo expression of Matrilysin (MMP-7) and Stromelysin-2 (MMP-10) in epithelial cells at the edges of intestinal ulcers52 suggests that these matrix metalloproteinases rather than MMP-1 and MMP-3 might be involved in the remodelling of the basement membrane and closure of intestinal ulcers.
In summary, our data suggest that increased expression of MMP-1 and MMP-3 is involved in the remodelling process of the ECM in inflamed colonic mucosa of IBD patients. Inhibitors of matrix metalloproteinases have been shown to effectively prevent tissue destruction in other inflammatory processes such as rheumatoid arthritis,53aphthous disease of the oral mucosa, and peridontal disease.54 Although our knowledge regarding the function of different matrix metalloproteinases in the intestinal tract is still limited, synthetic inhibitors specific for MMP-3 or MMP-1 warrant further exploration for their therapeutic efficacy in IBD.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grants La 1064 and Ro 674/10–1/2. The authors thank Mrs Heidrun Protz for technical assistance.
Abbreviations used in this paper
- MMP
- matrix metalloproteinase
- MT-MMP
- membrane-type MMP
- TIMP
- tissue inhibitor of matrix metalloproteinases
- PCR
- polymerase chain reaction
- IBD
- inflammatory bowel disease
- ECM
- extracellular matrix
- CD
- Crohn's disease
- UC
- ulcerative colitis
- mRNA
- messenger RNA
- cDNA
- complementary DNA
- APAAP
- alkaline phosphatase/anti-alkaline phosphatase
- RT
- reverse transcription
- PVDF
- polyvinyldifluoride
- ECL
- enhanced chemiluminescence