Article Text
Abstract
Objective Hepatocellular carcinoma (HCC) is a heterogeneous disease with poor prognosis and limited methods for predicting patient survival. The nature of the immune cells that infiltrate tumours is known to impact clinical outcome. However, the molecular events that regulate this infiltration require further understanding. Here the ability of immune genes expressed in the tumour microenvironment to predict disease progression was investigated.
Methods Using quantitative PCR, the expression of 14 immune genes in resected tumour tissues from 57 Singaporean patients was analysed. The nearest-template prediction method was used to derive and test a prognostic signature from this training cohort. The signature was then validated in an independent cohort of 98 patients from Hong Kong and Zurich. Intratumoural components expressing these critical immune genes were identified by in situ labelling. Regulation of these genes was analysed in vitro using the HCC cell line SNU-182.
Results The identified 14 immune-gene signature predicts patient survival in both the training cohort (p=0.0004 and HR=5.2) and the validation cohort (p=0.0051 and HR=2.5) irrespective of patient ethnicity and disease aetiology. Importantly, it predicts the survival of patients with early disease (stages I and II), for whom classical clinical parameters provide limited information. The lack of predictive power in late disease stages III and IV emphasises that a protective immune microenvironment has to be established early in order to impact disease progression significantly. This signature includes the chemokine genes CXCL10, CCL5 and CCL2, whose expression correlates with markers of T helper 1 (Th1), CD8+ T and natural killer (NK) cells. Inflammatory cytokines (tumour necrosis factor α, interferon γ) and Toll-like receptor 3 ligands stimulate intratumoural production of these chemokines which drive tumour infiltration by T and NK cells, leading to enhanced cancer cell death.
Conclusion A 14 immune-gene signature, which identifies molecular cues driving tumour infiltration by lymphocytes, accurately predicts survival of patients with HCC especially in early disease.
- Hepatocellular carcinoma
- immune-gene signature
- tumour microenvironment
- chemokines
- lymphocytes tumour infiltration
- immunology in hepatology
- tumour markers
- liver immunology
- immunohistochemistry
- acute hepatitis
- cancer
- oncogenes
- gastric cancer
- liver metastases
- pancreatic pathology
- pancreatic tumours
- molecular pathology
- gastrointestinal lymphoma
- liver biopsy
- surgical complications
- surgical oncology
- surgical resection
- immune response
This is an open-access article distributed under the terms of the Creative Commons Attribution Non-commercial License, which permits use, distribution, and reproduction in any medium, provided the original work is properly cited, the use is non commercial and is otherwise in compliance with the license. See: http://creativecommons.org/licenses/by-nc/2.0/ and http://creativecommons.org/licenses/by-nc/2.0/legalcode.
Statistics from Altmetric.com
- Hepatocellular carcinoma
- immune-gene signature
- tumour microenvironment
- chemokines
- lymphocytes tumour infiltration
- immunology in hepatology
- tumour markers
- liver immunology
- immunohistochemistry
- acute hepatitis
- cancer
- oncogenes
- gastric cancer
- liver metastases
- pancreatic pathology
- pancreatic tumours
- molecular pathology
- gastrointestinal lymphoma
- liver biopsy
- surgical complications
- surgical oncology
- surgical resection
- immune response
Significance of this study
What is already known about this subject?
Hepatocellular carcinoma (HCC) is a heterogeneous disease with poor prognosis and limited methods for predicting patient survival.
The nature of the immune cells that infiltrate tumours is known to impact clinical outcome.
In the past decade, several laboratories used gene expression profiling to define the molecular nature and identify prognostic signatures for HCC. However, little consensus was reached from such efforts and limited attention has so far been paid to the tumour immune microenvironment.
What are the new findings?
We identified a 14 immune-gene signature predictive of HCC patient survival in both the training cohort from Singapore (n=57; p=0.0004 and HR=5.2) and the validation cohort from Hong Kong and Zurich (n=98; p=0.0051 and HR=2.5) irrespective of patient ethnicity and disease aetiology.
The lack of predictive power in late-stage HCC shows that a protective immune microenvironment has to be established early in order to impact disease progression significantly.
Inflammatory cytokines (tumour necrosis factor α and interferon γ) and Toll-like receptor 3 ligands stimulate intratumoural production of chemokines especially CXCL10 and CCL5, which drive tumour infiltration by T and natural killer (NK) cells, hence leading to enhanced cancer cell death.
How might it impact on clinical practice in the foreseeable future?
The ability to make a prognosis in early stages of HCC will help in disease management such as in selection of patients with a better prognosis profile for liver transplantation.
Improved understanding of the molecular pathways leading to a protective local immune microenvironment will help in the rational design of new therapeutic approaches for patients with HCC.
Introduction
Hepatocellular carcinoma (HCC) claims >600 000 lives every year worldwide. HCC incidence is rising in Western countries partly due to increased hepatitis C virus infection.1 2 Limited treatments are available for patients with advanced disease. Curative resection remains the first line of treatment; however, due to a high recurrence rate, the overall survival of patients with HCC is poor. Sorafenib, a tyrosine kinase inhibitor recently approved for advanced HCC, brings only limited improvement in survival.3 More aggressive treatments, including liver transplantation for suitable patients, improve survival.4 However, identifying patients with HCC likely to benefit from such approaches remains challenging.
HCC is a heterogeneous disease comprising distinct molecular and clinical subgroups.5 6 This is largely due to the different HCC aetiologies which include hepatitis, and alcohol- and non-alcohol-induced cirrhosis. Geographical and ethnic variations further contribute to its heterogeneity.7 In the past decade, several laboratories used gene expression profiling to define the molecular nature and identify prognostic signatures for HCC.8–12 However, little consensus was reached from such efforts, illustrating the complexity and heterogeneity of this cancer. Each study focused on different molecular pathways, and limited attention has so far been paid to the tumour immune microenvironment.
It is now recognised that cancer progression is regulated by both cancer cell-intrinsic and microenvironmental factors. Among the latter, the nature and localisation of immune cells infiltrating the tumour play a central role. While tumour infiltration by myeloid cells is often associated with a poor prognosis,13 14 the presence of T helper 1 (Th1) or cytotoxic T cells correlates with a reduced risk of relapse in several cancers.15
We previously found that a proinflammatory tumour microenvironment correlates with prolonged survival in a cohort of Singaporean patients with HCC.16 In the current study, we identified a 14 immune-gene signature able to predict patient survival from this cohort and validated it in an independent cohort of patients from Hong Kong and Zurich. By combining transcriptome analysis, in situ labelling and in vitro experiments, we identified the cellular sources of the molecules corresponding to the gene signature. This approach revealed (1) a paracrine loop involving CXCL10, Toll-like receptor 3 (TLR3), tumour necrosis factor α (TNFα), and interferon γ (IFN-γ); and (2) an autocrine loop controlling CCL5 production. These two loops shape the immune milieu and recruit a potent antitumoral lymphoid infiltrate to the tumour of patients with longer survival. Our study shows that features derived from the tumour immune microenvironment are of general predictive value irrespective of HCC heterogeneity. Importantly, they determine the clinical outcome of patients with early-stage HCC for whom clinical parameters provide limited survival information. The lack of predictive power in late stages shows, for the first time in HCC, that the protective immune microenvironment has to be established early to promote long-term survival.
Materials and methods
Patients
One hundred and seventy-two resected HCC mRNA samples (one from each patient) were obtained from the National Cancer Centre (NCC), Singapore, Sg (n=61), the Queen Mary Hospital (QMH), Hong Kong, HK (n=56), and the University Hospital Zurich, Switzerland (n=55). All samples were obtained with Ethics Committee approval from patients who underwent curative resection from 1991 to 2009. After censoring patients with poor-quality gene expression profiles, data from Sg patients (n=57) were used as a training cohort to derive and test the survival prediction model, while HK (n=43) and Zurich (n=55) patients were used as an independent validation cohort. A total of 49 paraffin-embedded HCC samples (Sg, n=20; HK, n=23; Zurich, n=6) were obtained for immunohistochemistry or immunofluorescence labelling.
Clinical and demographic characteristics of the training and validation cohorts are summarised in table 1.
Comparison of clinical and demographic characteristics of patients with hepatocellular carcinoma in the training (Singapore) and validation (HK+Zurich) cohorts
Analysis of gene expression
Quantitative PCR (qPCR) analysis was performed on a total of 172 resected HCC mRNA samples. Primers were designed using Primer3 and qPCR was performed using iTaq SYBR Green Supermix with ROX (Bio-Rad Laboratories, Berkeley, California, USA), as described previously.16 Sixteen immune genes were selected for expression analysis, including the 11 previously published survival-related immune genes and five additional immune genes showing a strong trend of association with prolonged survival. Two of the genes, LTA and CCL22, were omitted from the gene list due to very low/undetectable expression in many of the validation cohorts. The relative gene expression level was calculated by normalisation to the housekeeping gene ACTB using MxPro software (Stratagene, Santa Clara, CA, USA).
Statistical analyses
Survival prediction was performed using the nearest-template prediction (NTP) method. The Cox score for each gene, which reflects the correlation between gene expression level and patient survival, was calculated as described previously.10 The prognosis prediction for each sample was made based on the proximity of its gene expression level to either of the templates of poor or good prognosis as defined by the vectors of weighted Cox scores. The survival predictor was evaluated in the training cohort (Sg, n=57) using a leave-one-out cross-validation, and tested on the independent validation cohort (HK, n=43 and Zurich, n=55). NTP was also validated by the bootstrap method as described previously.17 Two-class differential expression analysis was performed using GEPAS version 4.0 (http://gepas.bioinfo.cipf.es/).
Kaplan–Meier univariate survival analysis was performed using GraphPad Prism. Survival prediction is classified as ‘good prognosis’ or ‘poor prognosis’ according to the gene signature or as ‘low’ or ‘high’ as compared with the median of the relevant parameters. Patients who are still alive at last follow-up or are deceased due to causes unrelated to HCC were censored. Reported p values are obtained from the log-rank (Mantel–Cox) test.
Multivariate analysis by the Cox proportional hazards model was used to examine the gene signature in the context of clinical variables.
The NTP method and multivariate analyses were performed with the use of the R statistical package (http://www.r-project.org).
Immunohistochemistry (IHC) and immunofluorescence (IF)
IHC or IF labelling were performed on paraffin-embedded HCC samples as described before.16 The list of primary and secondary antibodies used is given in Supplementary table 1. IHC images were captured with an Olympus DP20 camera attached to a CX31 microscope. For IF, an Olympus FlourView FV1000 confocal microscope was used.
Quantification of positive cells was performed with ImagePro Software from 5–10 random fields at ×100 magnification for IHC, or 10–15 random fields at ×200 magnification for IF. The average value from all quantified fields was determined for each patient. Statistical analysis was performed with GraphPad Prism.
Isolation of tumour-infiltrating leukocytes (TILs)
Tumour tissues from patients with HCC (n=3) were obtained from Singapore General Hospital with Ethics Committee approval.
Tissues were homogenised using a Dispomix Drive (Xiril AG, Hombrechtikon, Switzerland). Tumour (T) and TILs were separated by a series of low speed centrifugations and filtration through a 100 μm filter (Millipore, Santa Clara, CA, USA) to remove large debris. A total of 1×106 cells were resuspended in Trizol (Invitrogen, Santa Clara, CA, USA) and RNA was converted to DNA using Taqman Reverse Transcriptase reagent (Applied Biosystems, Foster City, CA, USA) for qPCR analysis. Fraction purity assessed by flow cytometry was ∼90%.
In vitro chemokine production and transwell migration assays
The HCC cell line SNU-182 was obtained from the Korean Cell bank and cultured in complete RPMI medium. Cells were treated with 100 U/ml IFNγ (ImmunoTools, Friesoythe, Germany), 10 ng/ml TNFα, 50 μg/ml poly(I:C) (InvivoGen, SanDiego, CA, USA) or with a combination of IFNγ and TNFα, or IFNγ and poly(I:C). After 24 h, culture supernatants were collected for ELISA and cells were harvested for RNA isolation. RNA isolation, cDNA conversion and qPCR for CXCL10, CCL5 and CCL2 were performed as described above. ELISAs were performed to detect CXCL10, CCL5 and CCL2 using kits from R&D Systems (CXCL10 and CCL5; R&D Systems, Inc., Minneapolis, USA) and eBiosciences (CCL2; eBioscience, San Diego, CA, USA) according to the manufacturers' instructions. Absorbance intensity was analysed using a Tecan microplate reader.
For transwell migration assay, SNU-182 cells unstimulated or stimulated with IFNγ and poly(I:C) as described above were seeded into 24-well plates. After 24 h, 1×106 peripheral blood monocytes (PBMCs) from healthy donors (n=3) untreated or pretreated with anti-CXCR3 (25 μg/ml; clone 1C6, BD Pharmingen) or anti-CCR5 (10 μg/ml; clone 2D7, BD Pharmingen, San Diego, CA, USA) neutralising antibodies at 37°C for 1.5 h were added onto the transwell filter inserts (3 μm pore size, BD Falcon, San Diego, CA, USA). Transmigration was assessed after 3 h.
Results
Identification and validation of an immune-gene signature predicting overall survival of patients with HCC
We previously characterised the expression profile of 49 immune-related genes in 61 resected HCC tumour samples from Singapore, and found 11 immune genes whose expression was associated with superior patient survival.16 In the current study, we analysed the RNA expression of 14 immune genes: TNF, IL6, CCL2, NCR3, CCR2, TLR4, FCGR1A, CEACAM8, TLR3, CXCL10, CCL5, TBX21, CD8A and IFNG. We used NTP to identify and cross-validate (by the leave-one-out method) a 14 immune-gene signature predictive of overall survival in 57 Singaporean patients with resectable HCC (as a training cohort). The NTP method was chosen because it allows independent prediction for each sample and is less sensitive to differences in sample processing and analysis.18 The signature was then validated in an independent cohort of patients from Hong Kong (n=43) and Zurich (n=55) (figure 1A). Bootstrapping analysis also showed similar results (Supplementary figure 1).
Identification and validation of a 14 immune-gene signature predictive of overall survival in patients with hepatocellular carcinoma (HCC). (A) Study design for the identification of a 14 immune-gene signature derived from the training cohort (Sg, n=57) and validated in an independent cohort of patients from HK (n=43) and Zurich (n=55). NTP, nearest-template prediction; qPCR, quantitative PCR. (B and C) Heat maps showing the expression profile of the 14 immune genes (log values) in (B) the training cohort and (C) the validation cohort. Patients are classified as good or poor prognosis according to prediction by the immune-gene signature. FDR, p value of the t test adjusted for false discovery rate (multiple testing). Kaplan–Meier analyses for survival in (D) the training cohort, based on leave-one-out cross-validation testing and in (E) the independent validation cohort. Good and poor prognosis refers to the outcome predicted by the immune signature. p, log rank test p value.
In general, the 14 immune genes display higher expression in patients with good prognosis in both the training (figure 1B) and the validation cohort (figure 1C). The relative importance of each gene was assessed using its Cox score (Supplementary table 1). Despite the differences in patient ethnicity and disease stage (table 1), the herein presented 14-gene signature accurately predicts patient survival in both the training cohort (p=0.0004 and HR=5.2; figure 1D) and the validation cohort (p=0.0051 and HR=2.5; figure 1E). Multivariate analysis showed that this gene signature is an independent predictor of survival together with stage when six parameters are analysed (table 2). Strikingly, when stage IV patients were excluded, the immune signature was the only predictor of survival (table 2).
Multivariate analysis of the 14 immune-gene signature
Superior predictive power of the 14 immune-gene signature in early-stage patients
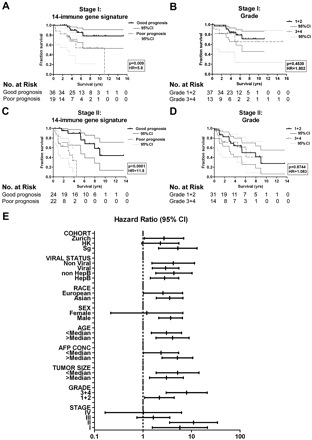
In the Singapore cohort, 60% of patients presented with stage I disease at diagnosis (table 1). We therefore measured the performance of the identified immune signature in patients with early (stages I and II) disease and compared it with clinical parameters generally used for prognosis of such patients. First, we noted that stage I (n=55) and II (n=46) patients (from both the training and validation cohorts) present a wide range of survival times, from a few months to >15 years (Supplementary figure 2). The immune signature accurately predicted the overall survival of these patients in Kaplan–Meier analyses (stage I, p=0.009, HR=5.8; stage II, p<0.0001, HR=11.8) (figure 2A,C). In contrast, clinical parameters such as grade (figure 2B,D), serum α-fetoprotein (AFP) concentration or tumour size (Supplementary figure 2) did not predict overall survival of these patients. Similar results were obtained from bootstrapping analysis (Supplementary figure 1).
Superior prognostic power of the 14 immune-gene signature compared with clinical parameters. Kaplan–Meier analyses for survival of (A) stage I patients (n=55, training and validation cohort) according to the immune-gene signature accurately predicts patient survival; (B) stage I patients according to grade (n=50); (C) stage II patients (n=46, training and validation cohort) according to the immune gene signature accurately predicts patient survival and (D) stage II patients according to grade (n=45). p, log rank test p value. (E) The plot shows HRs with 95% CI for subgroups of patients according to clinical and demographic characteristics. Age, median=61; AFP conc (α-fetoprotein concentration), median=20 ng/ml; tumour size, median=4.3 cm.
The predictive power of the 14-gene signature was also tested in various subgroups of patients (figure 2E). Interestingly, it did not predict the survival of stage III or IV patients. Therefore, the immune signature allows a robust and reliable prediction of overall survival in patients with early HCC for whom classical clinical parameters are not significant.
CXCL10, CCL5 and CCL2 expression correlates with intratumoural infiltration of Th1, CD8+ T and NK cells
Chemokine and chemokine receptor genes such as CXCL10, CCL5, CCL2 and CCR2 constitute a prominent group in the immune signature identified. Since chemokines are critical for attracting immune cells,19 we predicted that expression of these chemokines would correlate with tumour infiltration by defined immune cell subsets. To investigate this, we searched for correlations at the transcriptional level in 172 patient samples from both the training and validation cohorts. RNA expression of CXCL10, CCL5 and CCL2 correlated with markers of Th1 cells (TBX21), CD8+ T (CD8A) and NK (NCR3) cells (marked in red, figure 3A). Interestingly, TBX21, CD8A and NCR3 are also among the genes present in the signature. There was no correlation between expression of these chemokines and markers of other immune cell subsets such as macrophages (CD14 and CD68), Th2 (IL13), Th17 (IL17), Treg (FoxP3 and IL10), B (CD19) or dendritic (CD83) cells (marked in blue, figure 3A). This shows that CXCL10, CCL5 and CCL2 are associated with, and likely to attract specifically, Th1, CD8+ T and NK cells into HCC tumours.
CXCL10, CCL5 and CCL2 expression correlates with tumour infiltration by T and natural killer (NK) cells. (A) In patients with hepatocellular carcinoma (HCC; training and validation cohort, n=172), CXCL10, CCL5 and CCL2 RNA positively correlate with TBX21, CD8A and NCR3 (marked in red) but not with CD14, CD68, CD19, CD83, IL13, IL17, FOXP3 or IL10 (marked in blue). Graphs show p values against Pearson correlation coefficients r. The dotted line shows the limit of significance (p<0.05). (B) Representative immunofluorescence images showing higher density of CXCL10-expressing cells (red) in a tumour sample with high (left) versus low (right) density of infiltrating CD8+ and CD56+ cells as quantified by immunohistochemistry (IHC). The area in the rectangle is magnified in the left inset. Bar=50 μm; ×400 magnification. (C) Correlation of CXCL10 protein expression with the density of CD8+ (left) and CD56+ (right) immune cells. CXCL10 expression was determined by quantification of the CXCL10-labelled area, and CD8+ and CD56+ cell densities were measured by IHC in tumour fields of patient samples (CD8+, n=27; CD56+, n=19, training and validation cohort). p Values and correlation coefficients (r) were calculated using the Spearman correlation test. DAPI, 4′,6-diamidino-2-phenylindole.
To support this further, we measured the surface expression of CXCR3, CCR5 and CCR2 (the main receptors for CXCL10, CCL5 and CCL2, respectively) on PBMCs from healthy donors and patients with HCC, as well as on infiltrating leucocytes isolated from freshly resected tumours (TILs) or adjacent non-tumoural tissues (non-tumour-infiltrating lymphocytes or NILs). Flow cytometry analysis showed that T and NK cells represent the majority of the immune subsets expressing CXCR3 and CCR5 (Supplementary figure 3A). Furthermore, a greater percentage of T and NK cells express CCR5 and CCR2 in the PBMCs, TILs and NILs of patients as compared with healthy donor PBMCs (Supplementary figure 3A). This observation may indicate an increased propensity of T and NK cells from HCC patients to be attracted by CCL5 and CCL2.
We also analysed CXCL10 expression in tumour sections using IF. We first verified that CXCL10-specific IF correlated with mRNA expression (Supplementary figure 3B). We next showed that higher CXCL10-specific IF (figure 3B) was observed in samples with a higher density of CD8+ and CD56+ cells, as determined by IHC. Further quantification showed that the CXCL10 IF correlated with the density of CD8+ T cells and CD56+ NK cells (CD8, n=27, p=0.028, r=0.42; and CD56, n=19, p=0.042, r=0.47) (figure 3C) and also with patient survival (n=25, p=0.024, HR= 3.5) (Supplementary figure 3C).
Taken together, these data strongly suggest that CXCL10, CCL5 and CCL2 are the main chemokines attracting Th1 T cells, CD8+ T cells and NK cells into the tumour microenvironment.
Chemokines associated with patient survival are produced by both cancer cells and TILs
To understand the molecular interactions taking place within the tumour, we sought to identify the source of CXCL10, CCL5 and CCL2 within HCC. Single-cell suspensions from fresh tumour samples were separated into tumour cells and TILs, followed by chemokine expression analysis using qPCR. The three chemokine genes were transcribed in both tumour cells and TILs (figure 4A). Furthermore, when CXCL10 and CCL5 expression was analysed in situ by IHC, many chemokine-producing cells exhibited cancer cell morphology (figure 4B). CXCL10 was also expressed by TILs. IF on tumour sections, combining labelling for CXCL10 and immune cell markers (CD68, CD3 and CD20), revealed that most of the CXCL10-producing immune cells co-expressed CD68 (figure 4C) but not T or B cell markers (data not shown). Similarly, we found co-localisation of CCL5 and CD68 (figure 4D). Hence, macrophages within HCC tumours express both CXCL10 and CCL5.
CXCL10, CCL5 and CCL2 are produced by both immune and cancer cells within hepatocellular carcinoma (HCC) tumours. (A) Quantitative PCR (qPCR) analysis of CXCL10, CCL5 and CCL2 RNA expression in purified tumour cells (Tumour), tumour-infiltrating leucocytes (TIL) and unfractionated HCC nodules (HCC) from freshly resected tumours. The chemokines are expressed in all three compartments. Graphs show means and SD normalised to Tumour. (B) Representative immunohistochemistry images of CXCL10 (left) and CCL5 (right) showing expression in cells with cancer cell morphology. Bar=50 μm; ×200 magnification. (C) Representative immunofluorescence (IF) images showing co-localisation of CXCL10 (red) and CD68 (green). Bar=20 μm; ×800 magnification. (D) Representative IF images showing co-localisation of CCL5 (red) with either CD68 or CD3 (green). Bar=20 μm; ×800 magnification. DAPI, 4′,6-diamidino-2-phenylindole.
Besides macrophages, CCL5 was also produced by CD3+ T cells (figure 4D). Given the ability of CCL5 to attract T cells, this suggests an autocrine loop in which CCL5 produced by macrophages and/or cancer cells attracts T cells, which produce more CCL5 to amplify T cell infiltration further.
TNFα, IFNγ and TLR3 ligands induce expression of CXCL10, CCL5 and CCL2 by HCC cells and induce transmigration of T and NK cells
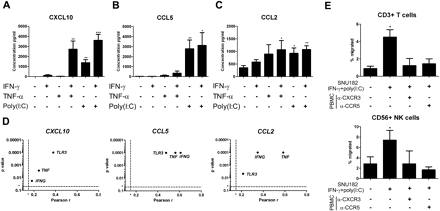
TNFα, IFNγ and TLR agonists stimulate CXCL10, CCL2 and CCL5 secretion by monocytes/macrophages,20–22 but little is known about the regulation of these chemokines in cancer cells. We used the HCC cell line SNU-182 to address this question. SNU-182 cells were treated with IFNγ, TNFα and the TLR3 ligand poly(I:C) separately or in combination, and culture supernatants were analysed. While IFNγ or TNFα alone had little effect, CXCL10 was strongly induced by the combination of IFNγ and TNFα (figure 5A). Poly(I:C) alone significantly induced CXCL10 expression, and this effect was further enhanced by addition of IFNγ (figure 5A). Poly(I:C) also induced CCL5 expression, while IFNγ or TNFα alone or in combination had no detectable effect (figure 5B). All three factors induced CCL2 expression, but no synergistic effect was observed (figure 5C). Chemokine gene induction could be observed by qPCR already 6 h after treatment (data not shown).
The production of CXCL10, CCL5 and CCL2 by HCC cell lines is induced by interferon γ (IFNγ), tumour necrosis factor α (TNFα) and Toll-like receptor 3 (TLR3) ligands. ELISA for (A) CXCL10, (B) CCL5 and (C) CCL2 concentration in culture supernatants from the SNU-182 hepatocellular carcinoma (HCC) cell line 24 h after stimulation with IFNγ, TNFα and/or poly(I:C). Two-tailed Student unpaired t test; *p<0.05; **p<0.01; ***p<0.001 compared with unstimulated control. Graphs show the means and SD from three independent experiments. (D) CXCL10, CCL5 and CCL2 RNA are positively correlated with IFNG, TNF and TLR3 in patients with HCC (training and validation cohort, n=172). Graphs show the p value against Pearson correlation coefficients r. Dotted lines show limits of significance for r (r=0.15) and p (p=0.05). (E) Transmigration assay with peripheral blood mononuclear cells (PBMCs) isolated from healthy donors (n=3) towards unstimulated or stimulated SNU-182 cells with IFNγ and poly(I:C) 24 h prior to transmigration. In blocking experiments, PBMCs were pretreated with anti-CXCR3 or anti-CCR5 neutralising antibodies at 37°C for 1.5 h. Graphs show means and SEM. p Values were calculated using paired t test against basal transmigration towards unstimulated HCC. *p<0.05. NK, natural killer.
To validate these observations in patient samples, we compared RNA expression of CXCL10, CCL5 and CCL2 and that of IFNG, TNF and TLR3 within tumours. Expression of the three chemokines correlated with that of IFNG, TNF and TLR3 (n=172 patients from both the training and validation cohorts; figure 5D).
A transwell migration assay was performed using stimulated SNU-182 cells and healthy donor PBMCs. The induction of chemokines in stimulated SNU-182 cells induced transmigration of T (fivefold increase) and NK cells (2.5-fold increase), without affecting other leucocytes (data not shown). Transmigration of T and NK cells was abolished when PBMCs were pretreated with anti-CXCR3 (CXCL10) or anti-CCR5 (CCL5) neutralising antibodies (figure 5E).
Taken together, these data indicate that IFNγ, TNFα and TLR3 ligands are potent inducers of the survival-associated chemokines CXCL10, CCL5 and CCL2. These chemokines attract T and NK cells which, upon activation, produce more IFNγ, triggering a paracrine loop leading to further amplification of chemokine production and lymphocyte infiltration.
Lymphocyte-attracting chemokines are associated with enhanced cancer cell death
CD8A and NCR3, two genes specific for CD8+ T cells and NK cells, respectively, are present in our gene signature and globally more expressed in long-term survivors. This is indeed reflected by enhanced infiltration of CD8+ T and CD56+ NK cells within the tumour samples from patients with longer survival (figure 6A, a subset of patients chosen for validation n=36 or 46). Kaplan–Meier analyses showed that a higher density of infiltrating CD8+ T (n=46, p<0.0001, HR=7.9) and CD56+ NK cells (n=36, p=0.016, HR=3.7) correlated with patient survival (figure 6B). Importantly, this was not observed for CD68+ macrophages (Supplementary figure 4). In this subset of patients, the current immune signature was superior at predicting patient survival compared with tumour infiltration by T cells or NK cells.
High chemokine expression levels, hence tumour infiltration by T and natural killer (NK) cells, are associated with superior patient survival. (A) Representative immunohistochemistry (IHC) images of CD8 and CD56 labelling (in red) showing higher density of CD8+ T and CD56+ NK cells in tumours from patients with longer survival (median survival >3.9 years). Bar=50 μm; ×200 magnification. (B) Kaplan–Meier analysis showing that high density of intratumoural CD8+ and CD56+ immune cells is associated with superior patient survival. A subset of patients was chosen for immune cell quantification by IHC (CD8, n=46, median=74 cells per field; CD56, n=36, median= 42 cells per field; training and validation cohort). p, log rank p value. (C) CXCL10 (n=26) immunofluorescence and (D) Toll-like receptor 3 (TLR3) (n=39) IHC staining area positively correlated with the density of activated caspase-3-positive tumour cells. r, Spearman (CXCL10) or Pearson (TLR3) correlation coefficient. (E) Downregulation of CXCL10, CCL5, CCL2 and TLR3 RNA expression in stages II–IV (n=114) compared with stage I patients with hepatocellular carcinoma (HCC) (n=57). Graphs show means and SEM. p Values were calculated using two-tailed Mann–Whitney U test. *p<0.05; **p<0.01; ***p<0.001. (F) Model showing that the inflammatory cytokines tumour necrosis factor α (TNFα), interferon γ (IFNγ) and Toll receptor-like (TLR) ligands stimulate cancer cells or macrophages to produce the key chemokines CXCL10, CCL5 and CCL2. These chemokines induce tumour infiltration by T helper 1 (Th1), CD8+ T and NK cells which induce cancer cell killing and tumour control. Positive feedback loops result from the production of IFNγ by activated T or NK cells that further enhance CXCL10 production (red arrow) and CCL5 by activated T cells that can attract more T cells (blue arrow).
We previously reported that the density of CD8+ T cells and CD56+ NK cells in HCC tumours correlates with cancer cell apoptosis detected by activated caspase-3 staining.16 Since CXCL10 and TLR3 activation play a major role in recruiting these cells, we examined if CXCL10 and TLR3 expression correlates with cancer cell apoptosis. Indeed, protein expression of CXCL10 (n=26, p=0.02, r=0.45; figure 6C) and TLR3 (n=39, p=0.04, r=0.33; figure 6D), an important inducer of CXCL10, CCL5 and CCL2, correlated with activated caspase-3 expression in cancer cells. Taken together, these correlations suggest a model in which chemokines expressed by cancer cells recruit lymphocytes that kill cancer cells, thereby contributing to prolonged patient survival. Such a model would predict that during the course of disease progression, cancer cells with reduced chemokines and TLR3 expression will be selected. Indeed, tumours from patients with more advanced HCC (stages II–IV; n=114) exhibit significantly lower RNA expression of CXCL10, CCL5, CCL2 and TLR3 than those from stage I patients (n=57) (figure 6E). This further confirms the crucial role of chemokines in shaping a protective immune environment early in disease development.
Discussion
In the present study we identified an immune signature which predicts the survival in resectable HCC irrespective of patient ethnicity or disease aetiology. Interestingly, it predicts the survival of early-stage patients for whom classical clinical parameters provide limited or no survival information. This signature, derived from resected HCC, comprises 14 genes coding for chemokines, inflammatory cytokines and lymphocyte markers. By combining transcriptome analysis, in situ staining and in vitro experiments, we identified regulatory circuits that shape and maintain a protective immune milieu within the tumour, leading to prolonged patient survival (figure 6F).
The immune signature was derived and tested using Singaporean patients and further validated in an independent cohort from Hong Kong and Zurich. The predictive value of the signature was also verified separately in various subgroups of patients (figure 2E). This consistency across different subsets of patients indicates that immune parameters determining disease progression are conserved irrespective of HCC heterogeneity. This is remarkable since HCC is known to be derived from multiple cell types (including hepatocytes or adult stem/progenitor cells)23 and caused by several aetiologies. Therefore, molecular features derived from the intratumoural immune response may be of better prognostic value than those relying on cancer cell characteristics. The loss of predictive power in female patients might be explained by the known gender disparity in the risk for HCC which is linked to oestrogen-mediated inhibition of interleukin 6 (IL-6)24 25 as IL6 is one of the genes in our signature.
Previously, several studies using genomic approaches identified gene signatures that stratify HCC patients according to clinical prognosis.8–12 These signatures were derived either from the adjacent non-tumour tissue or from the tumour itself. Signatures derived from the adjacent tissues emphasise risk factors for developing de novo tumours and support the ‘field defect’ hypothesis.10 Interestingly, immune characteristics of the adjacent liver tissues have also been shown to impact patient survival.9 10 On the other hand, signatures derived from the tumour itself focus on genes involved in proliferation and the cell cycle8 11 26 or on the identity of tumour-initiating cells.27 28 The current study is the first to focus exclusively on immune genes expressed within the tumour, and to show that the HCC immune milieu has an impact on disease outcome.
It may seem paradoxical that inflammation, an established risk factor for developing HCC, could play a protective role in HCC progression.29 30 For instance, IL-6 and TNFα were shown to promote HCC tumourigenesis.31–33 However, we found that these two cytokines correlate with longer patient survival in the present study. The beneficial impact of an active immune response within the tumour microenvironment is well established for non-small cell lung cancer (NSCLC),34 colorectal cancer35 36 and other malignancies.37 IL-6 and IL-8 were also reported to have a protective role in human colon adenomas.38 Similarly, depending on the mouse model, nuclear factor-κB (NF-κB), a major regulator of inflammation, suppresses or promotes HCC development.39 40 Additionally, expression of the same biomarker, for example, IL-6, in the serum or within the tumour may also reflect different biological processes.16 41 These apparent contradictions indicate that the effect of inflammation is context dependent and that the same cytokine may have opposite effects on HCC tumourigenesis and progression.42
In our model, inflammatory cytokines (TNFα and IFNγ) and TLR ligands (probably released from necrotic cells) induce chemokine expression within the tumour microenvironment. These chemokines (CXCL10, CCL5 and CCL2) could recruit immune cells, which display antitumour activity reflected by enhanced activated caspase-3 expression in cancer cells. Furthermore, infiltrating immune cells augment chemokine production (possibly through secretion of IFNγ or TNFα upon activation43) or directly secrete chemokines (CCL5), further stabilising the protective immune microenvironment. Such paracrine or autocrine loops are typical of complex biological systems as they provide efficient ways of amplifying signals and maintaining a particular immune status.44 Interestingly, no single cell type or molecular cue plays a unique role in shaping the immune microenvironment. Chemokines are produced by both cancer cells and TILs, while IFNγ is produced by Th1 and NK cells. Such redundancy also participates in the robustness of the protective environment, which has to be maintained for years in order to impact patient survival. The current immune signature predicts survival in patients in stages I and II but not in those in stages III and IV. This shows that a protective immune response has to be established early enough to be effective. Hence we propose that once the tumour has been established for prolonged periods of time, multiple layers of immune tolerance may prevent the efficacy of antitumour responses.45 46 It was therefore predictable and also shown in this study that cancer progression would be associated with downregulation of chemokines critically involved in the shaping of a protective immune microenvironment.
In summary, our study reveals extensive cross-talk between cancer cells and tumour-infiltrating immune cells in establishing a protective immune milieu able to delay HCC progression. Improved understanding of the molecular pathways leading to a protective immune microenvironment will help in the rational design of new therapeutic approaches for patients with HCC.
Acknowledgments
We wish to thank Professor Pierre-Alain Clavien from the University Hospital of Zurich for his provision of patient archives and information, Dr Peter Schraml for the provision of patient FFPE sections, Jay Tracy for preparation of RNA samples, and Yatanar Soe from the National Cancer Centre Singapore for co-ordinating the collection of fresh HCC tissues and patient blood samples. We thank Dr L. Robinson and J-L. Lebrun for manuscript editing, and B. Toh for help in figure preparation. IOLN is Loke Yew Professor in Pathology.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Files in this Data Supplement:
- Download Supplementary Data (PDF) - Manuscript file of format pdf
Footnotes
Funding This study was mainly funded by The Biomedical Sciences Institutes (BMSI), Agency for Science, Technology and Research (A*STAR), and partly funded by a Hong Kong Research Grants Council Collaborative Research Grant to IOLN (HKU 7/CRG/09).
Competing interests None.
Ethics approval This study was conducted with the approval of the respective Institutional Review Boards.
Provenance and peer review Not commissioned; externally peer reviewed.