Abstract
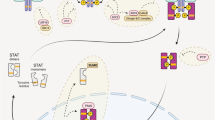
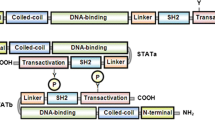
The transcription factor signal transducer and activator of transcription 3 (STAT3) is a critical regulator of multiple, diverse cellular processes. Heterozgyous, germline, loss-of-function mutations in STAT3 lead to the primary immune deficiency Hyper-IgE syndrome. Heterozygous, somatic, gain-of-function mutations in STAT3 have been reported in malignancy. Recently, germline, heterozygous mutations in STAT3 that confer a gain-of-function have been discovered and result in early-onset, multi-organ autoimmunity. This review summarizes what is known about the role of STAT3 in human disease.

Similar content being viewed by others
References
Zhong Z, Wen Z, Darnell Jr JE. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–8.
Levy DE, Darnell Jr JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–62.
Holland SM et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–19.
Minegishi Y et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–62.
Koskela HL et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–13.
Flanagan SE et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46(8):812–4.
Milner JD et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125(4):591–9.
Haapaniemi EM et al. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood. 2015;125(4):639–48.
O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368(2):161–70.
Mogensen TH. STAT3 and the Hyper-IgE syndrome: clinical presentation, genetic origin, pathogenesis, novel findings and remaining uncertainties. JAKSTAT. 2013;2(2), e23435.
UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12.
Ng IH et al. Selective STAT3-alpha or -beta expression reveals spliceform-specific phosphorylation kinetics, nuclear retention and distinct gene expression outcomes. Biochem J. 2012;447(1):125–36.
Shao H, Quintero AJ, Tweardy DJ. Identification and characterization of cis elements in the STAT3 gene regulating STAT3 alpha and STAT3 beta messenger RNA splicing. Blood. 2001;98(13):3853–6.
Dewilde S et al. Of alphas and betas: distinct and overlapping functions of STAT3 isoforms. Front Biosci. 2008;13:6501–14.
Maritano D et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5(4):401–9.
Chakraborty A, Tweardy DJ. Granulocyte colony-stimulating factor activates a 72-kDa isoform of STAT3 in human neutrophils. J Leukoc Biol. 1998;64(5):675–80.
Hevehan DL, Miller WM, Papoutsakis ET. Differential expression and phosphorylation of distinct STAT3 proteins during granulocytic differentiation. Blood. 2002;99(5):1627–37.
Villarino AV et al. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol. 2015;194(1):21–7.
Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36(4):515–28.
Wake, M.S. and C.J. Watson. STAT3 the oncogene - still eluding therapy? FEBS J. 2015;282(14):2600–11.
Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–51.
Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19(21):2628–37.
Carow B, Rottenberg ME. SOCS3, a major regulator of infection and inflammation. Front Immunol. 2014;5:58.
Wegrzyn J et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323(5915):793–7.
Camporeale A, Poli V. IL-6, IL-17 and STAT3: a holy trinity in auto-immunity? Front Biosci (Landmark Ed). 2012;17:2306–26.
Takeda K et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94(8):3801–4.
Tangye SG, Cook MC, Fulcher DA. Insights into the role of STAT3 in human lymphocyte differentiation as revealed by the hyper-IgE syndrome. J Immunol. 2009;182(1):21–8.
Kane A et al. STAT3 is a central regulator of lymphocyte differentiation and function. Curr Opin Immunol. 2014;28:49–57.
Fornek JL et al. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 2006;107(3):1085–91.
Cui W et al. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35(5):792–805.
Takeda K et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10(1):39–49.
Gotthardt D et al. Loss of STAT3 in murine NK cells enhances NK cell-dependent tumor surveillance. Blood. 2014;124(15):2370–9.
Davis SD, Schaller J, Wedgwood RJ. Job’s Syndrome. Recurrent, “cold”, staphylococcal abscesses. Lancet. 1966;1(7445):1013–5.
Buckley RH, Wray BB, Belmaker EZ. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics. 1972;49(1):59–70.
Grimbacher B et al. Hyper-IgE syndrome with recurrent infections--an autosomal dominant multisystem disorder. N Engl J Med. 1999;340(9):692–702.
Chandesris MO et al. Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore). 2012;91(4):e1–19.
Jiao H et al. Novel and recurrent STAT3 mutations in hyper-IgE syndrome patients from different ethnic groups. Mol Immunol. 2008;46(1):202–6.
Schimke LF et al. Diagnostic approach to the hyper-IgE syndromes: immunologic and clinical key findings to differentiate hyper-IgE syndromes from atopic dermatitis. J Allergy Clin Immunol. 2010;126(3):611–7.
Woellner C et al. Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J Allergy Clin Immunol. 2010;125(2):424–32.
Siegel AM et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35(5):806–18.
Chandesris MO et al. Frequent and widespread vascular abnormalities in human signal transducer and activator of transcription 3 deficiency. Circ Cardiovasc Genet. 2012;5(1):25–34.
Wallet N et al. Diffuse large B-cell lymphoma in hyperimmunoglobulinemia E syndrome. Clin Lymphoma Myeloma. 2007;7(6):425–7.
Sowerwine KJ, Holland SM, Freeman AF. Hyper-IgE syndrome update. Ann N Y Acad Sci. 2012;1250:25–32.
Siegel AM et al. Diminished allergic disease in patients with STAT3 mutations reveals a role for STAT3 signaling in mast cell degranulation. J Allergy Clin Immunol. 2013;132(6):1388–96.
Wolach O et al. Variable clinical expressivity of STAT3 mutation in hyperimmunoglobulin E syndrome: genetic and clinical studies of six patients. J Clin Immunol. 2014;34(2):163–70.
Grimbacher B et al. Genetic linkage of hyper-IgE syndrome to chromosome 4. Am J Hum Genet. 1999;65(3):735–44.
Milner JD et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–6.
Ma CS et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–7.
de Beaucoudrey L et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205(7):1543–50.
Burkett PR, Meyer Zu Horste G, Kuchroo VK. Pouring fuel on the fire: Th17 cells, the environment, and autoimmunity. J Clin Invest. 2015;125(6):2211–9.
Hsu AP et al. Intermediate phenotypes in patients with autosomal dominant hyper-IgE syndrome caused by somatic mosaicism. J Allergy Clin Immunol. 2013;131(6):1586–93.
Avery DT et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207(1):155–71.
Meyer-Bahlburg A et al. Heterozygous signal transducer and activator of transcription 3 mutations in hyper-IgE syndrome result in altered B-cell maturation. J Allergy Clin Immunol. 2012;129(2):559–62.
Deenick EK et al. Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J Exp Med. 2013;210(12):2739–53.
Ma CS et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119(17):3997–4008.
Ives ML et al. Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8(+) T-cell memory formation and function. J Allergy Clin Immunol. 2013;132(2):400–11.
Wilson RP et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. 2015;212(6):855–64.
Saito M et al. Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J Exp Med. 2011;208(2):235–49.
Steward-Tharp SM et al. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood. 2014;123(19):2978–87.
Zhang LY et al. Clinical features, STAT3 gene mutations and Th17 cell analysis in nine children with hyper-IgE syndrome in mainland China. Scand J Immunol. 2013;78(3):258–65.
Sundin M et al. Novel STAT3 mutation causing hyper-IgE syndrome: studies of the clinical course and immunopathology. J Clin Immunol. 2014;34(4):469–77.
Mogensen TH, Jakobsen MA, Larsen CS. Identification of a novel STAT3 mutation in a patient with hyper-IgE syndrome. Scand J Infect Dis. 2013;45(3):235–8.
Saikia B et al. Hyper-IgE syndrome with a novel STAT3 mutation-a single center study from India. Asian Pac J Allergy Immunol. 2014;32(4):321–7.
Renner ED et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122(1):181–7.
Yong PF et al. An update on the hyper-IgE syndromes. Arthritis Res Ther. 2012;14(6):228.
Goussetis E et al. Successful long-term immunologic reconstitution by allogeneic hematopoietic stem cell transplantation cures patients with autosomal dominant hyper-IgE syndrome. J Allergy Clin Immunol. 2010;126(2):392–4.
Nester TA et al. Effects of allogeneic peripheral stem cell transplantation in a patient with job syndrome of hyperimmunoglobulinemia E and recurrent infections. Am J Med. 1998;105(2):162–4.
Gennery AR et al. Bone marrow transplantation does not correct the hyper IgE syndrome. Bone Marrow Transplant. 2000;25(12):1303–5.
Patel NC et al. Successful haploidentical donor hematopoietic stem cell transplant and restoration of STAT3 function in an adolescent with autosomal dominant Hyper-IgE syndrome. J Clin Immunol. 2015;35(5):479–85.
Jerez A et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–57.
Rajala HL et al. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann Med. 2014;46(3):114–22.
Pilati C et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208(7):1359–66.
Jerez A et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122(14):2453–9.
Verbsky JW, Chatila TA. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and IPEX-related disorders: an evolving web of heritable autoimmune diseases. Curr Opin Pediatr. 2013;25(6):708–14.
Oliveira JB, Fleisher T. Autoimmune lymphoproliferative syndrome. Curr Opin Allergy Clin Immunol. 2004;4(6):497–503.
Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol. 2010;40(7):1830–5.
Chaudhry A et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–91.
Cohen AC et al. Cutting edge: decreased accumulation and regulatory function of CD4+ CD25(high) T cells in human STAT5b deficiency. J Immunol. 2006;177(5):2770–4.
Komatsu N et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med. 2014;20(1):62–8.
Gagliani N et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523(7559):221–5.
Tsoi LC et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44(12):1341–8.
Danoy P et al. Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn’s disease. PLoS Genet. 2010;6(12), e1001195.
Seddighzadeh M et al. Variants within STAT genes reveal association with anticitrullinated protein antibody-negative rheumatoid arthritis in 2 European populations. J Rheumatol. 2012;39(8):1509–16.
Barrett JC et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–62.
Frampton JE. Tocilizumab: a review of its use in the treatment of juvenile idiopathic arthritis. Paediatr Drugs. 2013;15(6):515–31.
van Vollenhoven RF et al. Tofacitinib or adalimumab versus placebo in rheumatoid arthritis. N Engl J Med. 2012;367(6):508–19.
Lee EB et al. Tofacitinib versus methotrexate in rheumatoid arthritis. N Engl J Med. 2014;370(25):2377–86.
Dupuis S et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293(5528):300–3.
Dupuis S et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–91.
Averbuch D et al. The clinical spectrum of patients with deficiency of signal transducer and activator of transcription-1. Pediatr Infect Dis J. 2011;30(4):352–5.
van de Veerdonk FL et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365(1):54–61.
Liu L et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208(8):1635–48.
Hambleton S et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110(8):3053–8.
Kofoed EM et al. Growth hormone insensitivity associated with a STAT5b mutation. N Engl J Med. 2003;349(12):1139–47.
Yildiz M et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668–79.
McDonald DR. TH17 deficiency in human disease. J Allergy Clin Immunol. 2012;129(6):1429–35.
He J et al. STAT3 mutations correlated with hyper-IgE syndrome lead to blockage of IL-6/STAT3 signalling pathway. J Biosci. 2012;37(2):243–57.
Uzel G et al. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol. 2013;131(6):1611–23.
Kristensen T et al. Clinical relevance of sensitive and quantitative STAT3 mutation analysis using next-generation sequencing in T-cell large granular lymphocytic leukemia. J Mol Diagn. 2014;16(4):382–92.
Ohgami RS et al. STAT3 mutations are frequent in CD30+ T-cell lymphomas and T-cell large granular lymphocytic leukemia. Leukemia. 2013;27(11):2244–7.
Miyazaki K et al. An adolescent with marked hyperimmuno-globulinemia E showing minimal change nephrotic syndrome and a STAT3 gene mutation. Clin Nephrol. 2011;75(4):369–73.
Giacomelli M et al. SH2-domain mutations in STAT3 in hyper-IgE syndrome patients result in impairment of IL-10 function. Eur J Immunol. 2011;41(10):3075–84.
Al Khatib S et al. Defects along the T(H)17 differentiation pathway underlie genetically distinct forms of the hyper IgE syndrome. J Allergy Clin Immunol. 2009;124(2):342–8.
Acknowledgments
The authors acknowledge the contributions of the members of the M.A.C. laboratory, particularly Ms. Nermina Saucier. This work was supported in part by the intramural research program of the National Institute of Allergy and Infectious Diseases, NIH. Work in M.A.C’s laboratory was supported by The Children’s Discovery Institute and St. Louis Children’s Hospital, The Scleroderma Foundation, the Rheumatic Diseases Core Center at Washington University (P30AR048335), and NIH training grant 5T32AR007279 (T.P.V.).
Conflict of Interest
T.P.V., J.D.M., and M.A.C. declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Up to 1.0 AMA PRA Category 1 Credit™ of Continuing Medical Education Credit can now be obtained by reading this review article and completing all activity components by visiting the Clinical Immunology Society web site at http://www.clinimmsoc.org/education/continuing-medical-education/e-learning-tools/journal-cme
Rights and permissions
About this article
Cite this article
Vogel, T.P., Milner, J.D. & Cooper, M.A. The Ying and Yang of STAT3 in Human Disease. J Clin Immunol 35, 615–623 (2015). https://doi.org/10.1007/s10875-015-0187-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-015-0187-8