Abstract
Tumor necrosis factor-α (TNF-α) is a cytokine that has an important role in immunity and inflammation by inducing cellular responses such as apoptosis. The transcription factor nuclear factor-κB (NF-κB) can paradoxically suppress and promote apoptosis in response to TNF-α. In this study, we found that p53 upregulated modulator of apoptosis (PUMA), a p53 downstream target and a BH3-only Bcl-2 family member, is directly regulated by NF-κB in response to TNF-α. TNF-α treatment led to increases in PUMA mRNA and protein levels in human colon cancer cells. The induction of PUMA was p53 independent, and mediated by the p65 component of NF-κB through a κB site in the PUMA promoter. The apoptotic effect of PUMA induction by TNF-α was unmasked by depleting the antiapoptotic protein Bcl-XL. In mice, PUMA was also induced by TNF-α in an NF-κB-dependent manner. TNF-α-induced apoptosis in a variety of tissues and cell types, including small intestinal epithelial cells, hepatocytes, and thymocytes, was markedly reduced in PUMA-deficient mice. Collectively, these results demonstrated that PUMA is a direct target of NF-κB and mediates TNF-α-induced apoptosis in vitro and in vivo.
Similar content being viewed by others
Main
Tumor necrosis factor-α (TNF-α) is a pleiotropic cytokine that has an important role in a variety of physiological and pathological processes such as immune and inflammatory responses.1 One of the most commonly observed activities of TNF-α is induction of apoptosis in different tissues and cell types. TNF-α-induced apoptosis is mediated by its interactions with cell-surface receptors such as TNF receptor 1 and TNF receptor 2 (extrinsic pathway), and mitochondrial dysfunction (intrinsic pathway).1 These two pathways cross talk through the actions of the BH3-only Bcl-2 family member Bid.2 Recent studies suggest the existence of multiple cross-talk mechanisms in TNF-α-induced apoptosis.3 Although a number of proapoptotic proteins can be induced by TNF-α,1 it has remained unclear how TNF-α triggers an apoptotic response.
Nuclear factor-κB (NF-κB) is the best-known mediator of TNF-α-associated cellular response. NF-κB is a group of dimeric transcription factors comprising members of the NF-κB/Rel family, including p50, p52, p65 (Rel-A), Rel-B, and c-Rel.4 Although p50 and p65 regulate the canonical NF-κB pathway, p52 and Rel-B are components of the noncanonical NF-κB pathway. The activity of NF-κB is normally kept in check by the IκB family of inhibitors, which bind to and sequester NF-κB in the cytoplasm.5 Activation of NF-κB is triggered by IκB phosphorylation by IKK kinases and subsequent proteasomal degradation, which allows NF-κB to translocate to the nucleus, where it binds to the κB consensus sequences and modulates numerous target genes.6
A large body of evidence has demonstrated a protective role of NF-κB in TNF-α-induced apoptosis in most tissues and cell types. For example, targeted deletion of p65 in mice leads to increased apoptosis in several tissues.7 The protection by NF-κB is due to transcriptional activation of a number of antiapoptotic proteins, such as c-FLIP, Bcl-2, Bcl-XL, cIAP2, and A1/Bfl-2.6 Cells without appropriate NF-κB activation can be eliminated by death-inducing signaling complex-mediated apoptosis through the extrinsic pathway.6 Conversely, NF-κB has been found to promote apoptosis under certain conditions by activating the expression of proapoptotic proteins, such as p53, Fas, Fas ligand, death receptor 4, and death receptor 5.8, 9 However, the mechanisms and physiological significance of NF-κB in apoptosis regulation remain controversial and poorly understood.10
p53 upregulated modulator of apoptosis (PUMA) is a downstream target of p53 and a BH3-only Bcl-2 family member.11, 12 It is induced by p53 following exposure to DNA-damaging agents, such as γ-irradiation and commonly used chemotherapeutic drugs.11, 12 It is also activated by a variety of nongenotoxic stimuli independent of p53, such as serum starvation, kinase inhibitors, glucocorticoids, endoplasmic reticulum stress, and ischemia/reperfusion.13, 14 The proapoptotic function of PUMA is mediated by its interactions with antiapoptotic Bcl-2 family members such as Bcl-2 and Bcl-XL,15 which lead to Bax/Bak-dependent mitochondrial dysfunction and caspase activation.16 Studies using PUMA-knockout (PUMA-KO) human somatic cells and mice revealed that PUMA is essential for DNA damage-induced and p53-dependent apoptosis.17, 18, 19 However, the mechanisms and functions of p53-independent PUMA induction remain unclear.
Because PUMA is involved in ischemia/reperfusion-induced intestinal injury,14 which can induce an inflammatory response, we investigated whether it is regulated by inflammatory cytokines. In this study, we demonstrated that PUMA is directly activated by NF-κB in response to TNF-α treatment in a p53-independent manner, and that PUMA is a critical mediator of TNF-α-induced apoptosis in vitro and in vivo.
Results
p53-independent induction of PUMA by TNF-α
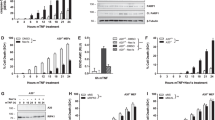
To determine the role of PUMA in TNF-α-induced apoptosis, HCT116 colon cancer cells were treated with 20 ng/ml of TNF-α. Both PUMA mRNA and protein were induced by TNF-α within several hours, with the peak level of PUMA mRNA induction at 12 h (Figure 1a), and that of protein at 24 h (Figure 1b). To determine whether p53 is involved in PUMA induction, we exposed isogenic HCT116 cells with a knockout of p53 (p53-KO) or a deletion of the p53-binding site in the PUMA promoter (BS-KO)20 to TNF-α. Induction of PUMA mRNA and protein was unaffected in these cells (Figure 1c and d). TNF-α treatment resulted in PUMA induction in six additional colon cancer cell lines regardless of their p53 status (Figure 1e). The induction of PUMA by TNF-α was also independent of FoxO3a (Supplementary Figure S1a), another transcription factor that can activate PUMA.21 Furthermore, several other BH3-only proteins, including Bad, Bim, and Noxa but not Bid, were induced by TNF-α in the parental and p53-KO HCT116 cells (Supplementary Figure S1b). These observations demonstrated that TNF-α induces the expression of BH3-only proteins including PUMA through p53-independent mechanisms.
p53-independent induction of PUMA by TNF-α in colon cancer cells. Colorectal cancer cells were treated with 20 ng/ml TNF-α. PUMA mRNA and protein at different time points after treatment were analyzed by RT-PCR and western blotting, respectively. (a) Time course of PUMA mRNA induction by TNF-α in HCT116 colon cancer cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was a control. (b) Time course of PUMA protein induction by TNF-α in HCT116 cells. Phospho-p65 (Ser536; P-p65) and α-tubulin were also analyzed. (c) Real-time RT-PCR analysis of PUMA mRNA expression in parental and p53-knockout (p53-KO) HCT116 cells following TNF-α treatment for 12 h. Values represent means±S.D. of three independent experiments. (d) Induction of PUMA protein by TNF-α treatment for 24 h in wild-type (WT), p53-KO, and p53-binding site-knockout (BS-KO) HCT116 cells. (e) Induction of PUMA protein in six colorectal cancer cell lines with different p53 status after TNF-α treatment for 24 h. Among these cell lines, DLD1, HT29, and SW480 contain mutant p53
p53-independent induction of PUMA by p65
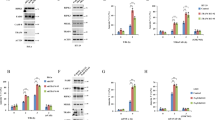
To identify the transcription factors that mediate PUMA induction in response to TNF-α, we subjected the DNA sequence of the PUMA promoter to bioinformatics analysis. Potential binding sites of several transcription factors known to mediate TNF-α response were identified, including those the NF-κB, ATF, IRF, and CREB families. Expression constructs of representative members of each family were transfected into cells (Supplementary Figure S2), and only the p65 subunit of NF-κB increased PUMA expression (Figure 2a). Transfection of p65 induced PUMA mRNA and protein in wild-type (WT), p53-KO, and BS-KO HCT116 cells (Figure 2b–d). Moreover, the level of phosphorylation of p65 at serine 536, an indicator of its transcriptional activity, was invariably correlated with PUMA induction in these cells (Figures 1b–e and 2d).
Induction of PUMA by the p65 subunit of NF-κB. PUMA mRNA and protein levels in cells at 24 h after transfection with a transcription factor or the control pcDNA vector were determined by RT-PCR and western blotting, respectively. (a) PUMA protein induction in p53-KO HCT116 cells by the indicated transcription factors. (b) PUMA mRNA induction in HCT116 cells by p65 transfection. (c) Real-time RT-PCR analysis of PUMA mRNA induction in parental and p53-KO HCT116 cells by p65 transfection. The results are means±S.D. of three independent experiments. (d) PUMA protein induction in WT, p53-KO, and BS-KO HCT116 cells by p65 transfection
p65 is necessary for PUMA induction by TNF-α
To determine whether NF-κB is necessary for the induction of PUMA by TNF-α, we first used BAY 11-7082, an inhibitor of the IKK kinases that are responsible for NF-κB activation. BAY 11-7082 pretreatment abrogated IκBα phosphorylation (S32/36) and degradation following TNF-α treatment (Supplementary Figure S3a), and suppressed subsequent PUMA induction and p65 nuclear translocation (Figure 3a). A nondegradable IκBα superrepressor mutant (S32/36A; IκBαM),22 which can specifically bind to and inhibit NF-κB, blocked PUMA induction by TNF-α and p65 nuclear translocation (Figure 3b). p65 siRNA but not the control siRNA abrogated PUMA induction by TNF-α in HCT116 cells (Figure 3c). Furthermore, PUMA induction by TNF-α in mouse embryonic fibroblast (MEF) cells was also found to be p65 dependent, but p53 independent (Figure 3d–f; Supplementary Figure S3b). The above data collectively indicate that p53-independent induction of PUMA by TNF-α is mediated by p65 through the canonical NF-κB pathway.
NF-κB is necessary for PUMA induction by TNF-α. (a) HCT116 cells were treated with TNF-α (20 ng/ml) in the presence or absence of the NF-κB inhibitor BAY 11-7082 (10 μM). Total cell lysates and nuclear extracts were isolated 24 h after treatment and analyzed by western blotting. Lamin A/C was a control for fractionation and loading. (b) HCT116 cells were transfected with the IκBα superrepressor mutant (S32/36A; IκBαM) or the control vector, and then treated by TNF-α (20 ng/ml) for 24 h. The indicated proteins were analyzed as in (a). (c) HCT116 cells were transfected with p65 or control scrambled siRNA, followed by TNF-α (20 ng/ml) treatment for 24 h and analysis of PUMA and p65 expression by western blotting. (d) WT and p65-knockout (p65-KO) MEF cells were treated with TNF-α (1 ng/ml) for 12 h and analyzed for PUMA mRNA expression by real-time RT-PCR. The results are means±S.D. of three independent experiments. (e) PUMA protein expression was analyzed by western blotting following the treatment as in (d). (f) PUMA protein expression in WT and p53-KO MEF cells was analyzed by western blotting after TNF-α (10 ng/ml) treatment for 18 h
p65 directly binds to the PUMA promoter to activate PUMA transcription in response to TNF-α
To determine whether p65 directly regulates PUMA expression at the transcriptional level, we constructed and analyzed a luciferase reporter construct containing the putative κB site in the PUMA promoter (Figure 4a). TNF-α treatment or co-transfection with p65 markedly activated the PUMA reporter (Figure 4b and c). Pretreating cells with the NF-κB inhibitor BAY 11-7082 blunted the activation of PUMA reporter by TNF-α (Figure 4d). Transfecting cells with the IκBα superrepressor mutant, but not the control pcDNA vector, also inhibited the activation of PUMA reporter (Figure 4e). Importantly, introducing mutations into the κB site abolished the responsiveness of the reporter to TNF-α treatment or p65 transfection (Figure 4a–c). Chromatin immunoprecipitation (ChIP) experiments showed that recruitment of p65 to the PUMA promoter region containing the κB site was greatly enhanced following TNF-α treatment for 6 h (Figure 4f). Together, these results indicate that p65 directly binds to the κB site in the PUMA promoter to drive its transcriptional activation in response to TNF-α treatment.
p65 directly activates PUMA transcription in response to TNF-α treatment. (a) Schematic representation of the PUMA genomic locus and reporter constructs. The sequences of WT and mutant κB sites are indicated. (b) WT and mutant PUMA reporter activities in HCT116 cells treated with TNF-α. The reporter activities were measured 24 h after TNF-α treatment, and normalized to those of β-galactosidase (relative luciferase activities, RLA). (c) WT and mutant PUMA reporter activities in HCT116 cells transfected with p65 or the control vector. The reporter activities were measured 24 h after transfection. (d) PUMA reporter activities in HCT116 cells treated with TNF-α for 24 h, with or without the NF-κB inhibitor BAY 11-7082 (10 μM). (e) PUMA reporter activities in HCT116 cells transfected with the IκBα superrepressor mutant (IκBαM) before TNF-α treatment for 24 h. (f) The binding of p65 to the PUMA promoter in HCT116 cells was analyzed by chromatin immunoprecipitation (ChIP) following TNF-α treatment for 12 h. ‘Input’ was equivalent to 1% of the starting materials for IP. IgG was used as a negative control. TNF-α was used at 20 ng/ml in (b)–(f). The results in (b)–(e) are means±S.D. of three independent experiments. Statistical analysis was performed using Student's t-test
PUMA-dependent apoptosis induced by TNF-α in colon cancer cells
Only low levels (<20%) of apoptosis were detected in colon cancer cells following TNF-α treatment. This might be due to simultaneous induction of the antiapoptotic NF-κB targets by TNF-α, such as Bcl-2 and Bcl-XL (Figure 5a), in addition to PUMA. Indeed, knockdown of Bcl-XL by siRNA, but not that of cIAP1 or Mcl-1, led to a significant increase in TNF-α-induced apoptosis (Figure 5b; Supplementary Figure S4a and S4b), consistent with the previous finding that Bcl-XL is the major survival factor in colon cancer cells.15 However, the effect of Bcl-XL knockdown on apoptosis induction was much reduced in the previously described PUMA-KO HCT116 cells (Figure 5b).17 Analysis of apoptosis using annexin V/propidium iodide (PI) staining confirmed this difference (Figure 5c). The critical function of PUMA is not cell line specific, as TNF-α-induced apoptosis following Bcl-XL knockdown was also significantly attenuated in the previously described PUMA-KO DLD1 colon cancer cells,20 compared with the WT DLD1 cells (Figure 5d). TNF-α-induced caspases 3, 8, and 9 activation; Bid cleavage; and release of cytochrome c were also suppressed in the PUMA-KO cells (Figure 5e and f), indicating that PUMA mediates TNF-α-induced apoptosis by promoting cytochrome c release and caspase activation through the mitochondrial pathway. We then analyzed FADD, an adapter protein that mediates TNF-α-induced apoptosis through the extrinsic pathway.1 Stable transfection of a dominant-negative (DN) FADD mutant slightly lowered TNF-α-induced apoptosis, but did not affect PUMA-dependent apoptosis following Bcl-XL knockdown (Figure 5g). The expression of DN FADD also left TNF-α-induced PUMA induction, p65 phosphorylation, and IκBα degradation intact (Supplementary Figure S4c). These results suggest that NF-κB-mediated PUMA induction represents a novel mechanism mediating TNF-α-induced apoptosis, and that overexpression of Bcl-XL can compromise TNF-α-induced and PUMA-mediated apoptosis.
TNF-α induced and PUMA-dependent apoptosis in colon cancer cells. (a) The indicated proteins in HCT116 cells treated with TNF-α for 24 h were analyzed by western blotting. (b) WT and PUMA-knockout (PUMA-KO) HCT116 cells were transfected with Bcl-XL or the control scrambled siRNA for 24 h, and then treated with TNF-α. PUMA and Bcl-XL expression was analyzed by western blotting following transfection (upper panel). Apoptosis was analyzed by nuclear staining after TNF-α treatment for 24 h (lower panel). (c) Following TNF-α treatment as in (b), cells were stained by annexin V/PI and analyzed by flow cytometry. The percentages of annexin V-positive (apoptotic) cells are within the two right quadrants. (d) WT and PUMA-KO DLD1 cells were analyzed for TNF-α-induced apoptosis following Bcl-XL knockdown as in (b). (e) Caspases 8, 3, and 9 activation and Bid cleavage in WT and PUMA-KO HCT116 cells following TNF-α treatment for 24 h. Arrowheads indicate caspase cleavage fragments. (f) After TNF-α treatment for 24 h, mitochondrial and cytosolic fractions were isolated from WT and PUMA-KO HCT116 cells and probed for cytochrome c by western blotting. Cytochrome oxidase subunit IV (Cox IV) and α-tubulin, which are expressed in the mitochondria and cytosol, respectively, were used as the controls for loading and fractionation. (g) A dominant-negative (DN) FADD mutant was transfected into WT and PUMA-KO HCT116 cells and stable cell lines were isolated. The expression of endogenous and DN FADD in the parental (Par) and two stable DN FADD-expressing cell lines (DN1 and DN2) was analyzed by western blotting (upper panel). Cells were transfected with Bcl-XL or scrambled siRNA and treated with TNF-α as in (b). Apoptosis was analyzed by nuclear staining after TNF-α treatment for 24 h (lower panel). TNF-α was used at 20 ng/ml in (a)–(g). The results in (b), (d), and (g) are means±S.D. of three independent experiments
PUMA is necessary for TNF-α-induced apoptosis in the intestinal epithelium
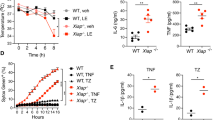
To determine the role of PUMA in TNF-α-induced apoptosis in vivo, we, on the basis of previous studies,23, 24 intravenously (i.v.) injected TNF-α at several doses (2, 4, and 10 μg; n=3) in WT and PUMA-KO mice. We first investigated whether PUMA contributes to TNF-α-induced apoptosis in the small intestine, which is susceptible to PUMA- or TNF-α-mediated tissue injury.14, 23 The expression of PUMA was elevated following TNF-α treatment (Figure 6a; Supplementary Figure S5a), which was inhibited by BAY 11-7082 (Figure 6b). In addition, other Bcl-2 family members, including Bcl-XL and Bim, were also induced by TNF-α in the small intestine (Supplementary Figure S5b). Dose-dependent apoptosis was observed in the crypts and villi of the WT mice at 8 h following TNF-α treatment (Figures 6c and d; Supplementary Figure S5c). In contrast, TNF-α-induced apoptosis was blocked to a large extent (60–90%) in the crypts and villus epithelium (P<0.001), but only slightly attenuated in the villus-inside of PUMA-KO mice (Figure 6c and d). TNF-α-induced caspase 3 activation was almost completely blocked in the crypts and villus epithelium of the PUMA-KO mice (Figure 6e; Supplementary Figure S5d).
PUMA mediates TNF-α-induced intestinal apoptosis. (a) PUMA expression in the small intestine of WT and PUMA-KO mice treated with TNF-α at indicated doses for 8 h was analyzed by western blotting. (b) WT mice were treated with TNF-α (2 μg) for 8 h with or without pretreatment with the NF-κB inhibitor BAY 11-7082 (200 μg). PUMA expression in the small intestine after TNF-α injection was analyzed by western blotting. (c) Representative pictures of TUNEL analysis of small intestine from WT and PUMA-KO mice treated with 2 μg of TNF-α for 8 h (original magnification × 400). (d) TUNEL-positive cells in crypts, villus epithelium, and villus-inside of small intestine after TNF-α treatment for 8 h in at least 20 randomly selected fields. (e) Active caspase 3-positive cells after TNF-α treatment for 8 h identified by IHC were counted in at least 20 randomly selected fields. Values are means±S.D., n=3 in each group in (d) and (e)
PUMA mediates TNF-α-induced hepatocyte apoptosis
We then extended the analysis to the liver, in which pathological conditions are often associated with deregulation of TNF-α-induced apoptosis. The expression of PUMA, but not that of other BH3-only proteins, was increased in the liver of WT mice following TNF-α treatment for 8 h (Figure 7a; Supplementary Figure S6a and S6b). Pretreating mice with the NF-κB inhibitor BAY 11-7082 attenuated the induction of PUMA by TNF-α (Figure 7b). TNF-α induced dose-dependent apoptosis in WT hepatocytes, but apoptosis was blocked by 50–70% in the PUMA-KO mice (Figure 7c and d). Furthermore, caspase 3 activation induced by TNF-α at different doses was abrogated in the hepatocytes of PUMA-KO mice (Figure 7e and f; Supplementary Figure S6c), indicating that PUMA mediates TNF-α-induced hepatocyte apoptosis. Analyses of additional organs revealed that the apoptotic response to TNF-α was also decreased in the colon, but not altered in the lung, of PUMA-KO mice (Supplementary Figure S7).
PUMA is required for TNF-α-induced hepatocyte apoptosis. (a) Liver extracts from WT mice treated with three doses of TNF-α were analyzed for PUMA expression by western blotting. β-Actin was the loading control. (b) WT mice with or without pretreatment with the NF-κB inhibitor BAY 11-7082 (200 μg) were treated with TNF-α (2 μg). Liver tissues were analyzed for PUMA expression by western blotting. (c) Representative pictures of TUNEL analysis of liver tissues after TNF-α (4 μg) treatment (original magnification × 400). (d) TUNEL-positive cells in liver sections from the WT and PUMA-KO mice treated with TNF-α were counted in at least 10 randomly selected fields. (e) Caspase 3 activation in liver tissues from WT and PUMA-KO mice treated with 4 μg of TNF-α was analyzed by western blotting. (f) Active caspase 3-positive hepatocytes identified by IHC were counted in at least 10 randomly selected fields. TNF-α treatment was for 8 h in (a)–(f). Values are means±S.D., n=3 in each group in (d) and (f)
TNF-α-induced and PUMA-dependent apoptosis in thymocytes
TNF-α can induce apoptosis in mouse thymocytes.25 TUNEL staining revealed more than two fold reduction in apoptosis in PUMA-KO thymocytes compared to WT ones (Figure 8a and b). Cell loss and apoptosis were also reduced in the spleens of PUMA-KO mice compared to WT mice (data not shown). To further investigate the role of PUMA in thymocyte apoptosis, we isolated primary thymocytes from WT and PUMA-KO mice and treated them with TNF-α at several concentrations (5–100 ng/ml). Both PUMA mRNA and protein were induced in WT thymocytes following TNF-α treatment (Figure 8c and d). WT thymocytes in culture underwent spontaneous apoptosis, which is partially PUMA dependent (Supplementary Figure S8). TNF-α treatment induced apoptosis in WT thymocytes, but had little effect on PUMA-KO thymocytes (Figure 8e), indicating that TNF-α-induced apoptosis in thymocytes is PUMA dependent.
PUMA is necessary for TNF-α-induced thymocyte apoptosis. (a) Thymocyte apoptosis was analyzed by TUNEL (brown) staining of thymic tissues collected at 8 h after TNF-α injection. Representative pictures from mice treated by 4 μg of TNF-α are shown (original magnification × 400). (b) Thymocyte apoptosis in (a) was quantified by dividing the number of TUNEL-positive cells by the number of counted cells in at least 10 randomly selected fields. Values are means±S.D., n=3 in each group. (c) Primary thymocytes were isolated from WT mice and treated with TNF-α (50 ng/ml). PUMA mRNA expression was analyzed by real-time RT-PCR after treatment for 8 or 18 h. (d) PUMA protein expression in primary thymocytes treated with TNF-α for 18 h from (c) was analyzed by western blotting. α-Tubulin was the loading control. (e) Induced cell death, which was calculated by subtracting the fraction of cells undergoing spontaneous apoptosis, in primary thymocytes isolated from WT and PUMA-KO mice was analyzed by annexin V/PI staining followed by flow cytometry after TNF-α treatment at indicated concentrations for 18 h. Values are means±S.D. of three independent experiments
Discussion
In this study, we identified PUMA as a novel target of NF-κB and a critical mediator of TNF-α-induced apoptosis in vitro and in vivo. PUMA mRNA and protein were consistently activated in cells treated with TNF-α. The induction of PUMA by TNF-α required the p65 subunit of NF-κB, and was mediated by a κB site located in the distal region of the PUMA promoter. These results established the first case of direct regulation of a BH3-only Bcl-2 family member by NF-κB in the TNF-α response. Previous studies have shown that PUMA can be induced by p53 in response to DNA-damaging agents, such as γ-irradiation and commonly used chemotherapeutic drugs,20 by the p53 homologue p73 in response to serum starvation,26 and by the transcription factor FoxO3a in response to cytokine/growth factor withdrawal.21 Our study revealed a new PUMA induction mechanism, which may be important in apoptosis triggered by inflammatory cytokines.
It was unexpected that NF-κB-mediated PUMA induction by TNF-α was completely p53 independent, considering the well-established cross talk between p53 and NF-κB in regulating gene expression.27 The p52 subunit, which is a component of the noncanonical NF-κB pathway, was found to cooperate with p53 in regulating p53 target genes including PUMA.28 However, the regulation of these genes by p52 seemed to suppress, rather than promote cell death.28 A recent study showed that phosphorylation of CREB-binding protein (CBP) by IKKα suppresses p53-mediated gene expression by switching the binding preference of CBP from p53 to NF-κB.29 Such a mechanism may be engaged in directing p65, instead of p53, to the PUMA promoter in response to TNF-α treatment. Several studies have shown that in some circumstances p53 and p65 can cooperatively induce apoptosis.30, 31 Because PUMA is one of the most potent inducers of apoptosis,11 such cooperative effects may result from coordinated induction of PUMA by p53 and p65. Activation of NF-κB is known to render cancer cells resistant to anticancer drugs. Inhibition of NF-κB has been explored as an attractive approach for anticancer therapies.32 However, our data suggest that NF-κB inhibition can compromise PUMA induction by inflammatory cytokines, which may be involved in tumor suppression and be beneficial for anticancer therapies.
PUMA is necessary for TNF-α-induced apoptosis as it functions as a novel link between the extrinsic and intrinsic apoptotic pathways. On activation by NF-κB through cell death receptors, PUMA promotes caspase activation and cytochrome c release. Among the BH3-only Bcl-2 family members, only Bid has been shown to have similar activity. In most circumstances, TNF-α is not an effective inducer of apoptosis in cultured cells, likely due to simultaneous induction of both proapoptotic and antiapoptotic proteins by NF-κB. Studies of TNF-α-induced apoptosis often rely on using transcription or translation inhibitors, which nonspecifically inhibit gene expression and often complicate data interpretation. We found that TNF-α simultaneously induced PUMA and the antiapoptotic proteins Bcl-2 and Bcl-XL in colon cancer cells. The apoptotic effect of NF-κB-mediated PUMA induction could be unmasked by inhibition of Bcl-XL, which is a potent survival factor and one of the commonly overexpressed genes in colorectal cancer.15 Previous studies found that NF-κB can induce both antiapoptotic and proapoptotic genes in neuronal cells,33 implying that the paradoxical activity of NF-κB in apoptosis is physiological, and is likely to be prevalent. Bcl-XL overexpression can neutralize the proapoptotic activity of PUMA, which may allow preneoplastic cells to evade the killing effects of inflammatory cytokines.
TNF-α administration into mice induced PUMA expression and PUMA-dependent apoptosis in tissues including small intestine and liver. Induction of PUMA by TNF-α in these tissues could be inhibited by the NF-κB inhibitor BAY 11-7082, suggesting that NF-κB can promote TNF-α-induced apoptosis by activating PUMA in these tissues. However, the induction of PUMA by TNF-α in vivo may involve more complicated mechanisms than that in colon cancer cells, and may engage other transcription factors. Whether PUMA induction in these tissues requires NF-κB needs to be further examined using more physiological systems including genetic models. In small intestine, PUMA was required for TNF-α-induced apoptosis in villus epithelial cells but not in villus-inside cells, which is similar to γ-irradiation-induced apoptosis reported in our recent study.34 Intestinal mucosal inflammation, which is initiated and perpetuated by cytokines in particular TNF-α, is associated with pathological conditions such as intestinal epithelial damage and inflammatory bowel disease. As a mediator of TNF-α-induced apoptosis, PUMA may contribute to inflammation-induced apoptosis and tissue damage, which is supported by our recent finding that PUMA is necessary for small intestinal apoptosis and tissue injury caused by ischemia/reperfusion, an inducer of inflammatory response.14 In contrast to our hepatocyte findings, a number of previous studies indicate a protective role of NF-κB in TNF-α-induced hepatocyte apoptosis.35 This difference may be explained by the doses of TNF-α (2–10 μg) used in our study, which were intended for studying intestinal apoptosis,23, 24 but higher than those normally used for inducing liver apoptosis. TNF-α at the doses we used may cause liver injury and an acute inflammatory response that triggers PUMA-dependent apoptosis. It would be interesting to further explore the connection between inflammation and tissue injury using PUMA-deficient mice.
PUMA deficiency suppressed TNF-α-induced apoptosis in the thymus, an essential immune response organ where T cells are produced and activated. NF-κB can promote thymocyte and T-cell apoptosis, which is critical for shaping the T-cell repertoire during thymocyte ontogeny. Our data suggest that the apoptotic effect of NF-κB in thymocytes is in part mediated by PUMA. The PUMA-dependent thymocyte apoptosis we observed may also involve other biological molecules induced by TNF-α, such as glucocorticoids. It has been shown that the glucocorticoid dexamethasone induces PUMA-dependent apoptosis in mouse thymocytes.18, 19 Studies from other groups showed a critical role of PUMA in thymocyte and lymphocyte apoptosis in different settings. For example, PUMA is required for thymocyte apoptosis induced by γ-irradiation and growth factor deprivation.18 Total thymocyte number is significantly increased in mice deficient in both PUMA and Bim.36 PUMA can also complement the function of Bim in controlling T-cell apoptosis in the termination of immune response.37
In summary, we demonstrated that PUMA is a direct target of NF-κB and a critical mediator of TNF-α-induced apoptosis in vitro and in vivo. Our data suggest that PUMA functions as a novel link between the extrinsic and intrinsic apoptotic pathways.
Materials and Methods
Cell culture and drug treatment
The human colorectal cancer cell lines, including HCT116, RKO, LOVO, Lim2405, which contain WT p53, and HT29, DLD1, and SW480, which contain mutant p53,11 were obtained from American Type Culture Collection (Manassas, VA, USA). p53-KO HCT116 cells were previously described,11 and were obtained from Dr. Bert Vogelstein (Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins). p53-binding site knockout (BS-KO) HCT116 cells were previously described.20 Immortalized WT, p53-KO, and p65-knockout (p65-KO) MEF cells were previously described.38 Colon cancer cells were maintained in McCoy's 5A medium (Invitrogen, Carlsbad, CA, USA). MEF cells were cultured using Dulbecco's modified Eagle's medium (BioWhittaker, Walkersville, MD, USA). All cell culture media were supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA), 100 U/ml of penicillin, and 100 mg/ml of streptomycin. Cells were maintained at 37°C in 5% CO2 atmosphere.
Cells were plated in 12-well plates at 20–30% density 24 h before treatment. Human TNF-α (R&D system, Minneapolis, MN, USA) was diluted with appropriate cell culture media. In some experiments, cells were pretreated with 10 μM of the NF-κB inhibitor BAY 11-7082 (Calbiochem, San Diego, CA, USA) 1 h before addition of TNF-α.
Transfection and reporter assays
Transfection was performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. siRNA transfection was performed 24 h before TNF-α treatment. siRNA duplexes, including FoxO3a siRNA (ON-TARGETplus siRNA J-003007-10), the previously described p65 siRNA,39 Bcl-XL siRNA,15 and the control scrambled siRNA, were from Dharmacon (Chicago, IL, USA).
PUMA luciferase reporter construct was generated by cloning a genomic fragment (Frag D; base 1470–2192 5′ to exon 1a) containing a κB site in the PUMA promoter into the pBV-Luc plasmid as previously described.26 Mutations were introduced into the κB site using QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). For reporter assays, cells were transfected with WT or mutant PUMA reporter along with the transfection control β-galactosidase reporter pCMVβ (Promega, Madison, WI, USA). Cell lysates were collected and luciferase activities were measured as previously described.11 All reporter experiments were performed in triplicate and repeated at least three times. p65 expression construct was obtained from Dr. David Derse at the National Cancer Institute, Frederick, MD, USA. IκBα superrepressor mutant (S32/36A; IκBαM) was previously described.22 The DN FADD construct was previously described,40 and obtained from Dr. Shi-Yong Sun at Emory University. Stable HCT116 and PUMA-KO cell lines expressing DN FADD were established by G418 (0.4 mg/ml) selection following DN FADD transfection as previously described.15 Expression vectors of several transcription factors, including CREB, IRF1, IRF2, IRF3, ATF1, ATF2, and ATF3, were constructed by cloning full-length cDNA fragments generated from reverse transcriptase (RT)-PCR into pcDNA3.1 vector. The inserts were verified by sequencing. The primer sequences are available on request.
Western blotting and antibodies
Western blotting was performed as previously described.20 Antibodies included those against human PUMA,17 murine PUMA (Abcam, Cambridge, MA, USA), human p53 (DO-1), Mcl-1, p65, IκBα and Lamin A/C (Santa Cruz Biotechnology, Santa Cruz, CA, USA), human phospho-p65 (Ser536), phospho-IκBα (Ser32/36), active caspase 3, caspase 8, and caspase 9 (Cell Signaling Technology, Danvers, MA, USA), cIAP1 (R&D Systems), Bim and Noxa (EMD Biosciences, Gibbstown, NJ, USA), Bad, FADD, and α-tubulin (BD Biosciences, San Diego, CA, USA). The Bid antibodies were a gift from Dr. Xiao-Ming Yin at University of Pittsburgh.
Reverse transcriptase-PCR and chromatin immunoprecipitation
Total RNA was isolated from treated cells using the RNAgents Total RNA Isolation System (Promega). First-strand cDNA was synthesized from 10 μg of total RNA using Superscript II reverse transcriptase (Invitrogen). PUMA cDNA was amplified using the primers 5′-CGACCTCAACGCACAGTACGA-3′, and 5′- AGGCACCTAATTGGGCTCCAT -3′. Real-time RT-PCR was performed in an MJ Mini Personal thermocycler (Bio-Rad, Hercules, CA, USA) using the same PUMA primers with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal control. The cycle conditions are available on request.
ChIP was performed using the Chromatin Immunoprecipitation Assay kit (Upstate Biotechnology, Lake Placid, NY, USA) as previously described.20 The precipitates were analyzed by PCR using primers 5′- CATGTAAGTGATGTCATATGTC -3′ and 5′- CTTCCTGGTCTTTTCCAAACT-3′.
Analysis of apoptosis
After treatment, cells were collected at different time points. For analysis of apoptosis by nuclear staining, cells were resuspended and fixed in PBS solution containing 3.7% formaldehyde, 0.5% Nonidet P-40, and 10 μg/ml Hoechst 33258 (Invitrogen). Apoptosis was assessed through microscopic visualization of condensed chromatin and micronucleation as described.16 For analysis of apoptosis by annexin V/PI staining, cells were stained by annexin Alexa 488 (Invitrogen) according to the manufacturer's instructions, counterstained by PI for cell nuclei, and then analyzed by flow cytometry as described.20 For analysis of cytochrome c release, mitochondrial and cytosolic fractions were isolated by the differential centrifugation method previously described,20 and probed by western blotting for cytochrome c.
Animal experiments
The procedures of all animal experiments were approved by the Institutional Animal Care and Use Committee at University of Pittsburgh. Weight-matched and 6 to 8 week-old WT (PUMA+/+) and PUMA-KO (PUMA−/−) littermates on C57BL/6 background were generated by heterozygote breeding and identified by PCR genotyping as previously described.14, 18, 34 The mice were housed in micro isolator cages and allowed access to water and chow ad libitum.
Mice were i.v. injected with 2, 4, or 10 μg of recombinant murine TNF-α (ProSpec-Tany TechnoGene, Rehovot, Israel) in 400 μl PBS. The control animals were injected with PBS. In some experiments, mice were injected with 200 μg of the NF-κB inhibitor BAY 11-7082 (Calbiochem) 1 h before TNF-α injection. Mice were killed at 8 h after TNF-α injection. Three mice were used in each group.
TUNEL and immunostaining
After killing the animals, tissues were removed and rinsed with ice-cold PBS, and fixed in 10% neutral buffered formalin. The fixed tissues were embedded in paraffin and sectioned. Histological analysis was performed by hematoxylin and eosin staining. TUNEL staining was performed using ApopTag kit (Chemicon International, Temecula, CA, USA) according to the manufacturer's instructions. For thymocytes, the apoptotic index (%) was scored as the percentage of TUNEL-positive cells. For hepatocytes, apoptosis was scored by counting TUNEL-positive cells in at least 10 randomly selected fields. For small intestine, apoptosis was scored for crypt, villus epithelium, and villus-inside by counting TUNEL-positive cells in 20 randomly selected crypts or villi.
Active caspase 3 immunohistochemistry (IHC) was performed as previously described,34 with an active caspase 3 antibody (Cell Signaling Technology) at 1 : 100 dilution at room temperature for 2 h. The signals were detected with ABC and DAB kits (Vector Laboratory, Burlingame, CA, USA). The sections were counterstained with hematoxylin.
Bioinformatics and statistical analyses
Transcription factor binding sites in the PUMA promoter region were identified using the TF Search Program (http://mbs.cbrc.jp/research/db/TFSEARCH). Statistical analysis was performed using GraphPad Prism IV software (GraphPad Software, La Jolla, CA, USA). Data were analyzed by unpaired t-test or ANOVA in which multiple comparisons were performed using the least-significant difference method. P-values <0.05 were considered to be statistically significant. The means±one standard deviation (S.D.) are displayed in the figures.
Abbreviations
- TNF-α:
-
tumor necrosis factor-α
- PUMA:
-
p53 upregulated modulator of apoptosis
- CBP:
-
CREB-binding protein
- MEF:
-
mouse embryonic fibroblast
References
Wajant H, Pfizenmaier K, Scheurich P . Tumor necrosis factor signaling. Cell Death Differ 2003; 10: 45–65.
Danial NN, Korsmeyer SJ . Cell death. Critical control points. Cell 2004; 116: 205–219.
Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 2007; 12: 445–456.
Hoffmann A, Baltimore D . Circuitry of nuclear factor kappaB signaling. Immunol Rev 2006; 210: 171–186.
Gilmore TD . Introduction to NF-kappaB: players, pathways, perspectives. Oncogene 2006; 25: 6680–6684.
Karin M, Lin A . NF-kappaB at the crossroads of life and death. Nat Immunol 2002; 3: 221–227.
Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D . Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 1995; 376: 167–170.
Burstein E, Duckett CS . Dying for NF-kappaB? Control of cell death by transcriptional regulation of the apoptotic machinery. Curr Opin Cell Biol 2003; 15: 732–737.
Dutta J, Fan Y, Gupta N, Fan G, Gelinas C . Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene 2006; 25: 6800–6816.
Perkins ND, Gilmore TD . Good cop, bad cop: the different faces of NF-kappaB. Cell Death Differ 2006; 13: 759–772.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B . PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001; 7: 673–682.
Nakano K, Vousden KH . PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694.
Yu J, Zhang L . No PUMA, no death: implications for p53-dependent apoptosis. Cancer Cell 2003; 4: 248–249.
Wu B, Qiu W, Wang P, Yu H, Cheng T, Zambetti GP et al. p53 independent induction of PUMA mediates intestinal apoptosis in response to ischaemia-reperfusion. Gut 2007; 56: 645–654.
Ming L, Wang P, Bank A, Yu J, Zhang L . PUMA dissociates Bax and BCL-XL to induce apoptosis in colon cancer cells. J Biol Chem 2006; 281: 16034–16042.
Yu J, Wang P, Ming L, Wood MA, Zhang L . SMAC/Diablo mediates the proapoptotic function of PUMA by regulating PUMA-induced mitochondrial events. Oncogene 2007; 26: 4189–4198.
Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L . PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA 2003; 100: 1931–1936.
Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003; 4: 321–328.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science 2003; 302: 1036–1038.
Wang P, Yu J, Zhang L . The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci USA 2007; 104: 4054–4059.
You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med 2006; 203: 1657–1663.
Hu J, Nakano H, Sakurai H, Colburn NH . Insufficient p65 phosphorylation at S536 specifically contributes to the lack of NF-kappaB activation and transformation in resistant JB6 cells. Carcinogenesis 2004; 25: 1991–2003.
Haimovitz-Friedman A, Cordon-Cardo C, Bayoumy S, Garzotto M, McLoughlin M, Gallily R et al. Lipopolysaccharide induces disseminated endothelial apoptosis requiring ceramide generation. J Exp Med 1997; 186: 1831–1841.
Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 2000; 289: 2350–2354.
Hernandez-Caselles T, Stutman O . Immune functions of tumor necrosis factor. I. Tumor necrosis factor induces apoptosis of mouse thymocytes and can also stimulate or inhibit IL-6-induced proliferation depending on the concentration of mitogenic costimulation. J Immunol 1993; 151: 3999–4012.
Ming L, Sakaida T, Yue W, Jha A, Zhang L, Yu J . Sp1 and p73 activate PUMA following serum starvation. Carcinogenesis 2008; 29: 1878–1884.
Perkins ND . Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 2007; 8: 49–62.
Schumm K, Rocha S, Caamano J, Perkins ND . Regulation of p53 tumour suppressor target gene expression by the p52 NF-kappaB subunit. EMBO J 2006; 25: 4820–4832.
Huang WC, Ju TK, Hung MC, Chen CC . Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol Cell 2007; 26: 75–87.
Ryan KM, Ernst MK, Rice NR, Vousden KH . Role of NF-kappaB in p53-mediated programmed cell death. Nature 2000; 404: 892–897.
Fujioka S, Schmidt C, Sclabas GM, Li Z, Pelicano H, Peng B et al. Stabilization of p53 is a novel mechanism for proapoptotic function of NF-kappaB. J Biol Chem 2004; 279: 27549–27559.
Gilmore TD, Herscovitch M . Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene 2006; 25: 6887–6899.
Mattson MP, Camandola S . NF-kappaB in neuronal plasticity and neurodegenerative disorders. J Clin Invest 2001; 107: 247–254.
Qiu W, Carson-Walter EB, Liu H, Epperly M, Greenberger JS, Zambetti GP et al. PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell 2008; 2: 576–583.
Ding WX, Yin XM . Dissection of the multiple mechanisms of TNF-alpha-induced apoptosis in liver injury. J Cell Mol Med 2004; 8: 445–454.
Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med 2006; 203: 2939–2951.
Fischer SF, Belz GT, Strasser A . BH3-only protein Puma contributes to death of antigen-specific T cells during shutdown of an immune response to acute viral infection. Proc Natl Acad Sci USA 2008; 105: 3035–3040.
Song L, Li J, Zhang D, Liu ZG, Ye J, Zhan Q et al. IKKbeta programs to turn on the GADD45alpha-MKK4-JNK apoptotic cascade specifically via p50 NF-kappaB in arsenite response. J Cell Biol 2006; 175: 607–617.
Braun T, Carvalho G, Coquelle A, Vozenin MC, Lepelley P, Hirsch F et al. NF-kappaB constitutes a potential therapeutic target in high-risk myelodysplastic syndrome. Blood 2006; 107: 1156–1165.
Liu X, Yue P, Zhou Z, Khuri FR, Sun SY . Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst 2004; 96: 1769–1780.
Acknowledgements
We thank Dr. Bert Vogelstein, Dr. Jing Hu, Dr. Shi-Yong Sun, Dr. Xiao-Ming Yin, and Dr. David Derse for reagents, and Dr. Sean Garrison for reading the paper. This work was supported by NIH grants CA106348, CA121105, American Cancer Society grant RSG-07-156-01-CNE, the Career Investigator Award form the American Lung Association/CHEST Foundation (LZ), the Flight Attendant Medical Research Institute (FAMRI), and the Alliance for Cancer Gene Therapy (ACGT), and NIH grant CA129829 (JY). LZ is a scholar of the V Foundation for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by J Silke
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, P., Qiu, W., Dudgeon, C. et al. PUMA is directly activated by NF-κB and contributes to TNF-α-induced apoptosis. Cell Death Differ 16, 1192–1202 (2009). https://doi.org/10.1038/cdd.2009.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.51
Keywords
This article is cited by
-
Silencing suppressor of cytokine signaling 3 induces apoptosis and activates the p-STAT3/NF-κB pathway in hypoxic cultivated H9c2 cells
Journal of Physiology and Biochemistry (2024)
-
Vitamin D and biomarkers of inflammation and oxidative stress among pregnant women: a systematic review of observational studies
BMC Immunology (2023)
-
Cooperative effects of RIG-I-like receptor signaling and IRF1 on DNA damage-induced cell death
Cell Death & Disease (2022)
-
MY11 exerts antitumor effects through activation of the NF-κB/PUMA signaling pathway in breast cancer
Investigational New Drugs (2022)
-
BRD9 inhibition promotes PUMA-dependent apoptosis and augments the effect of imatinib in gastrointestinal stromal tumors
Cell Death & Disease (2021)