Abstract
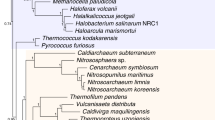
The ancestors of fungi are believed to be simple aquatic forms with flagellated spores, similar to members of the extant phylum Chytridiomycota (chytrids). Current classifications assume that chytrids form an early-diverging clade within the kingdom Fungi and imply a single loss of the spore flagellum, leading to the diversification of terrestrial fungi. Here we develop phylogenetic hypotheses for Fungi using data from six gene regions and nearly 200 species. Our results indicate that there may have been at least four independent losses of the flagellum in the kingdom Fungi. These losses of swimming spores coincided with the evolution of new mechanisms of spore dispersal, such as aerial dispersal in mycelial groups and polar tube eversion in the microsporidia (unicellular forms that lack mitochondria). The enigmatic microsporidia seem to be derived from an endoparasitic chytrid ancestor similar to Rozella allomycis, on the earliest diverging branch of the fungal phylogenetic tree.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Karol, K. G., McCourt, R. M., Cimino, M. T. & Delwiche, C. F. The closest living relatives of land plants. Science 294, 2351–2353 (2001)
Groth-Malonek, M., Pruchner, D., Grewe, F. & Knoop, V. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol. Biol. Evol. 22, 117–125 (2005)
Lang, B. F., O’Kelly, C., Nerad, T., Gray, M. W. & Burger, G. The closest unicellular relatives of animals. Curr. Biol. 12, 1773–1778 (2002)
James, T. Y., Porter, D., Leander, C. A., Vilgalys, R. & Longcore, J. E. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 78, 336–350 (2000)
Tanabe, Y., Saikawa, M., Watanabe, M. M. & Sugiyama, J. Molecular phylogeny of Zygomycota based on EF-1α and RPB1 sequences: limitations and utility of alternative markers to rDNA. Mol. Phylogenet. Evol. 30, 438–449 (2004)
Schüßler, A., Schwarzott, D. & Walker, C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421 (2001)
Tehler, A., Little, D. P. & Farris, J. S. The full-length phylogenetic tree from 1551 ribosomal sequences of chitinous fungi, Fungi. Mycol. Res. 107, 901–916 (2003)
Cavalier-Smith, T. & Chao, E. E. The opalozoan Apusomonas is related to the common ancestor of animals, fungi, and choanoflagellates. Proc. R. Soc. Lond. B 261, 1–9 (1995)
Steenkamp, E. T., Wright, J. & Baldauf, S. L. The protistan origins of animals and fungi. Mol. Biol. Evol. 23, 93–106 (2005)
Barr, D. J. S. Evolution and kingdoms of organisms from the perspective of a mycologist. Mycologia 84, 1–11 (1992)
Ustinova, I., Krienitz, L. & Huss, V. A. R. Hyaloraphidium curvatum is not a green alga, but a lower fungus; Amoebidium parasiticum is not a fungus, but a member of the DRIPs. Protist 151, 253–262 (2000)
Nagahama, T., Sato, H., Shimazu, M. & Sugiyama, J. Phylogenetic divergence of the entomophthoralean fungi: evidence from nuclear 18S ribosomal RNA gene sequences. Mycologia 87, 203–209 (1995)
McKerracher, L. J. & Heath, I. B. The structure and cycle of the nucleus-associated organelle in two species of Basidiobolus.. Mycologia 77, 412–417 (1985)
Blackwell, M. & Malloch, D. Similarity of Amphoromorpha and secondary capilliconidia of Basidiobolus.. Mycologia 81, 735–741 (1989)
Keeling, P. J. & Fast, N. M. in Insect–Fungal Associations: Ecology and Evolution (eds Vega, F. E. & Blackwell, M.) 97–118 (Oxford Univ. Press, Oxford, 2005)
Hirt, R. P. et al. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl Acad. Sci. USA 96, 580–585 (1999)
Katinka, M. D. et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi.. Nature 414, 450–453 (2001)
Keeling, P. J. Congruent evidence from α-tubulin and β-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet. Biol. 38, 298–309 (2003)
Cavalier-Smith, T. in The Mycota (eds McLaughlin, D. J., McLaughlin, E. G. & Lemke, P. A.) 3–37 (Springer, New York, 2001)
Corradi, N., Hijri, M., Fumagalli, L. & Sanders, I. R. Arbuscular mycorrhizal fungi (Glomeromycota) harbour ancient fungal tubulin genes that resemble those of the chytrids (Chytridiomycota). Fungal Genet. Biol. 41, 1037–1045 (2004)
Held, A. A. Rozella and Rozellopsis: Naked endoparasitic fungi which dress up as their hosts. Bot. Rev. 47, 451–515 (1981)
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002)
Gill, E. E. & Fast, N. M. Assessing the microsporidia–fungi relationship: combined phylogenetic analysis of eight genes. Gene 375, 103–109 (2006)
KirkP. M.CannonP. F.DavidJ. C.StalpersJ. A. (eds) Ainsworth & Bisby’s; Dictionary of the Fungi 60 (CAB International, Wallingford, UK, 2001)
Lutzoni, F. et al. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am. J. Bot. 91, 1446–1480 (2004)
Binder, M. et al. The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Systematics and Biodiversity 3, 113–157 (2005)
Moncalvo, J-M. et al. One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol. 23, 357–400 (2002)
Hibbett, D. S., Gilbert, L-B. & Donoghue, M. J. Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Nature 407, 506–508 (2000)
Medina, M. et al. Phylogeny of Opisthokonta and the evolution of multicellularity and complexity in Fungi and Metazoa. Int. J. Astrobiol. 2, 203–211 (2003)
Powell, M. J. Fine structure of the unwalled thallus of Rozella polyphagi in its host Polyphagus euglenae.. Mycologia 76, 1039–1048 (1984)
Berbee, M. L. & Taylor, J. W. in The Mycota (eds McLaughlin, D. J., McLaughlin, E. G. & Lemke, P. A.) 229–245 (Springer, New York, 2001)
Heckman, D. S. et al. Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129–1133 (2001)
Pirozynski, K. A. & Malloch, D. W. The origin of land plants: a matter of mycotrophism. Biosystems 5, 153–164 (1975)
Schüßler, A. Molecular phylogeny, taxonomy, and evolution of arbuscular mycorrhiza fungi and Geosiphon pyriformis.. Plant Soil 244, 75–83 (2002)
Keeling, P. J. & Inagaki, Y. A class of eukaryotic GTPase with a punctuate distribution suggesting multiple functional replacements of translation elongation factor 1α. Proc. Natl Acad. Sci. USA 101, 15380–15384 (2004)
Maddison, D. & Maddison, W. MacClade Version 4.05: Analysis of Phylogeny and Character Evolution (Sinauer Associates, Sunderland, Massachusetts, USA, 2002)
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003)
Huelsenbeck, J. P. & Ronquist, F. MrBayes: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001)
Wilgenbusch, J. C., Warren, D. L. & Swofford, D. L. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. http://ceb.csit.fsu.edu/awty (2004)
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002)
Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 1.0. http://mesquiteproject.org (2003)
Acknowledgements
Funding for the project was provided by the National Science Foundation’s ‘Assembling the Tree of Life’ and ‘Research Coordination Network’ programs. For technical assistance we thank L. Bukovnik, C. Roberts and H. Matthews. We also thank the following individuals for sharing research materials: M. C. Aime, W. R. Buck, M. S. Cole, P. Crane, Y. Dalpe, D. M. Hillis, S. L. Joneson, R. Petersen, C. Printzen, E. Vellinga, H. Whisler and A. Zavarzin. We are very thankful to B. Mueller, J. Harer, B. Rankin, J. Pormann and S. Dilda for providing access to the Duke CSEM computer cluster. P. Keeling provided unpublished information used to analyse EFL in Fungi.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Data for this project have been deposited in GenBank (see Supplementary Notes 1 for accession numbers), and the alignments can be accessed on the Assembling the Fungal Tree of Life website at http://www.aftol.org/.Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes 1
A table of the strains and species used and the sources and GenBank numbers for the gene sequences used in this study. (PDF 618 kb)
Supplementary Notes 2
These file shows the results of a Bayesian phylogenetic analysis of the six gene super-matrix using only nucleotide data. Note: Figure 1 of the manuscript shows a phylogeny based on a heterogeneous nucleotide-amino acid model, whereas this phylogeny uses nucleotides divided into genes and codons. (PDF 120 kb)
Supplementary Notes 3
This file contains the results of analysis of individual gene partitions (and relevant combinations) and the Bayesian posterior probabilities and maximum likelihood bootstrap support for nodes of interest. (PDF 66 kb)
Supplementary Notes 4
This file contains a phylogeny of a paralogous copy of elongation factor 1-α (EFL). It shows the species of basal fungi from which this gene copy has been sequenced, and demonstrates a relationship between Basidiobolus and the Entomophthorales. (PDF 115 kb)
Supplementary Notes 5
This file contains the methods and results from tests of long branch attraction between Rozella allomycis and microsporidia. (PDF 67 kb)
Rights and permissions
About this article
Cite this article
James, T., Kauff, F., Schoch, C. et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822 (2006). https://doi.org/10.1038/nature05110
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05110
This article is cited by
-
Plastiphily is linked to generic virulence traits of important human pathogenic fungi
Communications Earth & Environment (2024)
-
The first two mitochondrial genomes from Apiotrichum reveal mitochondrial evolution and different taxonomic assignment of Trichosporonales
IMA Fungus (2023)
-
Assessment of fungal spores and spore-like diversity in environmental samples by targeted lysis
BMC Microbiology (2023)
-
A fungal plant pathogen discovered in the Devonian Rhynie Chert
Nature Communications (2023)
-
Geographic population structure of the honeybee microsporidian parasite Vairimorpha (Nosema) ceranae in the South West Indian Ocean
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.