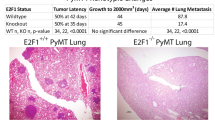
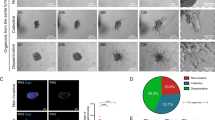
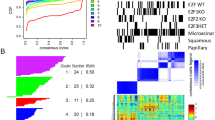
Abstract
Metastasis is a complex multistep process, which requires the concerted action of many genes and is the primary cause of cancer death. Both pathways that regulate metastasis enhancement and those that regulate its suppression contribute to the tumour dissemination process. To identify new metastasis suppressors, we set up a forward genetic screen in a mouse model. We transduced a genome-wide RNA interference (RNAi) library into the non-metastatic 168FARN breast cancer cell line and orthotopically transplanted the cells into mouse mammary fat pads. We then selected cells that could metastasize to the lung and identified an RNAi for the KLF17 gene. Conversely, we demonstrate that ectopic expression of KLF17 in a highly metastatic 4T1 breast cancer cell line inhibits the ability of cells to metastasize from the mammary fat pad to the lung. We also show that suppression of KLF17 expression promotes breast cancer cell invasion and epithelial–mesenchymal transition (EMT), and that KLF17 protein functions by directly binding to the promoter region of Id1 (which encodes a key metastasis regulator in breast cancer) to inhibit its transcription. Finally, we demonstrate that KLF17 expression is significantly downregulated in primary human breast cancer samples and that the combined expression pattern of KLF17 and Id1 can serve as a potential biomarker for lymph node metastasis in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Gupta, G. P. & Massague, J. Cancer Metastasis: building a framework. Cell 127, 679 (2006).
Steeg, P. S. Tumor metastasis: mechanistic insights and clinical challenges. Nature Rev. Med. 12, 895–904 (2006).
Fidler, I. J. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nature Rev. Cancer 3, 1–6 (2003).
Steeg, P. S. Metastasis suppressors alter the signal transduction of cancer cells. Nature Rev. Cancer 3, 55–63 (2003).
Yang, J. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Muller, A. et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 (2001).
Eccles, S. A. & Welch, D. R. Metastasis: recent discoveries and novel treatment strategies. Lancet 369, 1742–1757 (2007).
Chambers, A. F., Groom, A. C. & MacDonald, I. C. Dissemination and growth of cancer cells in metastatic sites. Nature Rev. Cancer 2, 563–572 (2002).
Stafford, L. J., Vaidya, K. S. & Welch, D. R. Metastasis suppressor genes in cancer. Int. J. Biochem. Cell Biol. 40, 874–891 (2008).
Yoshida, B. A., Sokoloff, M. M., Welch, D. R. & Rinker-Schaeffer, C. W. Metastasis-suppressor genes: a review and perspective on an emerging field. J. Natl Cancer Inst. 92, 1717–1730 (2000).
Grimm, S. The art and design of genetic screens: mammalian culture cells. Nature Rev. Genet. 5, 179–189 (2004).
Schlabach, M. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008).
Dasgupta, R., Kaykas, A., Moon, R. T. & Perrimon, N. Functional genomic analysis of the Wnt-Wingless signaling pathway. Science 308, 826–832 (2005).
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systemic RNA interference. Nature 408, 325–330 (2000).
Paddison, P. J. et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427–431 (2004).
Berns K et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431–7 (2004).
Kolfschoten, I. G. et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity Cell 121, 849–58 (2005).
Aslakson, C. J. & Miller, F. R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52, 1399–1405 (1992).
Aslakson, C. J., Rak, J. W., Miller, B. E. & Miller, F. R. Differential influence of organ site on three subpopulations of a single mouse mammary tumor at two distinct steps in metastasis. Int. J. Cancer 47, 466–472 (1991).
Miller, F., Jones, R. F., Jacob, J., Kong, Y. C., Wei, Y. Z. From breast cancer immunology to her-2 DNA vaccine and autoimmune sequelae. Breast Dis. 20, 43–51 (2004).
Dexter, D. L. et al. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 38, 3174–3181 (1978).
Gumireddy, K. et al. An in vivo selection for metastasis promoting genes in the mouse. Proc. Natl Acad. Sci. USA 104, 6696–6701 (2007).
Lomberk, G., Urrutia, R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392, 1–11 (2005).
Kaczynski, J., Cook, T., Urrutia, R. Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206 (2003).
Vliet, J. V. et al. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics 87, 474–482 (2005).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Nuez, B., Michalovich, D., Bygrave, A., Ploemacher, R., Grosveld, F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375, 316–318 (1995).
Perkins, A. C., Sharpe, A. H. & Orkin, S. H. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375, 318–322 (1995).
Kuo, C. T., Veselits, M. L., Leiden, J. M. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science 277, 1986–1990 (1997).
Foster, K. W. et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 60, 6488–6495 (2000).
Narla, G. et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294, 2563–2566 (2001).
Ghaleb, A. M. et al. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 15, 92–96 (2005).
Yan, W., Burns, K. H., Ma, L., Matzuk, M. M. Identification of Zfp393, a germ-cell specific gene encoding a novel zinc finger protein. Mech. of Dev. 118, 233–239 (2002).
Hugo, H. et al. Epithelial-mesenchymal and mesenchymal-epithelial transition in carcinoma progression. J. Cell. Physiol. 213, 374–383 (2007).
Huber, M. A., Kraut, N. & Beug, H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17, 548–558 (2005).
Zavadil, J., Cermak, L., Soto-Nieves, N., Bottinger, E. P. Integration of TGF-β/Smad and Jagged1/Notch signaling in endothelial to mesenchymal transition. EMBO J. 23, 1155–1165 (2004).
Liebner, S. et al. β-catinin is required for endothelial-mesenchymal transformation furing heart cushion development in the mouse. J. Cell. Biol. 166, 359–367 (2004).
Moody, S. E. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005).
Hartwell, K. A. et al. The spemann organizer gene, goosecoid, promotes tumor metastasis. Proc. Natl. Acad. Sci. USA 103, 18969–18974 (2006).
Ruzinova, M. B. & Benezra, R. Id proteins in development, cell cycle and cancer. Trends in Cell Biol. 13, 410–418 (2003).
Sikder, H. A., Devlin, M. K., Dunlap, S., Ryu, B. & Alani, R. Id proteins in cell growth and tumorigenesis Cancer Cell 3, 525–530 (2003).
Ivarone, A. & Lasorella, A. ID proteins as targets in cancer and tools in neurobiology. Trends in Mol. Med. 12, 588–594 (2006).
Ying, Q., Nichols, J., Chambers, I. & Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 (2003).
Li, Y., Yang, J., Luo, J., Dedhar, S., Liu, Y. Tubular epithelial cell dedifferentiation is driven by the helix-loop-helix transcriptional inhibitor Id1. J. Am. Soc. Nephrol. 18, 449–460 (2007).
Lyden, D. et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumor xenograft. Nature 401, 670–677 (1999).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Swarbrick, A., Roy, E., Allen, T. & Bishop, J. M. Id1 cooperates with oncogenic Ras to induce metastastic mammary carcinoma by subversion of the cellular senescence response. Proc. Natl. Acad. Sci. USA 105, 5402–5407 (2008).
Schoppmann, S. F. et al. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int. J. Cancer 104, 677–682 (2003).
Lin, C. Q. et al. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 60, 1332–1340 (2000).
Gupta, G. P. et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc. Natl. Acad. Sci. USA 104, 19506–19511 (2007).
Henke, E. et al. Peptide-conjugated antise oligonucleotides for targeted inhibition of a transcription regulator in vivo. Nature Biotechnol. 26, 91–100 (2008).
McAllister, S. D., Christian, R. T., Horowitz, M. P., Garcia, A. & Desprez, P. Cannabidiol as a novel inhibitor of Id-1 gene expression in aggressive breast cancer cells. Mol. Cancer Ther. 6, 2921–2927 (2007).
Kataoka, M. et al. An agent that increases tumor suppressor transgene product coupled with systemic transgene delivery inhibits growth of metastatic lung cancer in vivo. Cancer Res. 58, 4761–4765 (1998).
Huang, Q. et al. The microRNAs miR-373 and miR-520c promote tumor invasion and metastasis. Nature Cell Biol. 10, 202–210 (2008).
Mukhopadhyay, A., Deplancke, B., Walhout, A. J. M., Tissenbaum, H. A. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real- time PCR to study transcription factors binding to DNA in Caenorhabditis elegans. Nature Protoc. 3, 698–709 (2008).
Acknowledgements
We would like to thank F. Miller for providing 168FARN and 4T1 cells, R. Weinberg for providing the HMEL cell line, J. Price for providing the MDA-MB-435 cell line, C. Chang and W. Horng for assistance with microarray analysis and J. Hayden and F. Keeney for assistance with microscopy. Q.H. is supported by the Breast Cancer Alliance, Pardee Foundation, V Foundation. Q.H. and L.C.S. are supported by the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (PA DOH; P30 CA10815) and L.C.S. is supported by a PA DOH grant (SAP 4100020718). G.C. and L.Z. are supported by NCI ovarian SPORE (P50-CA83638), Ovarian Cancer Research Fund and Mary Kay Ash Charitable Foundation.
Author information
Authors and Affiliations
Contributions
K.G. and Q. H. designed the experiments. K.G and A.L. performed the experiments. P.A.G. performed statistical analysis. A.J.K. performed pathological analysis. D.K., G.C. and L.Z. provided experimental materials. K.G., A.L., L.C.S and Q.H. analysed data. K.G., L.C.S. and Q.H. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1747 kb)
Rights and permissions
About this article
Cite this article
Gumireddy, K., Li, A., Gimotty, P. et al. KLF17 is a negative regulator of epithelial–mesenchymal transition and metastasis in breast cancer. Nat Cell Biol 11, 1297–1304 (2009). https://doi.org/10.1038/ncb1974
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1974
This article is cited by
-
ID2 promotes tumor progression and metastasis in thyroid cancer
Endocrine (2024)
-
KLF17 promotes human naive pluripotency through repressing MAPK3 and ZIC2
Science China Life Sciences (2022)
-
Exploring liver cancer biology through functional genetic screens
Nature Reviews Gastroenterology & Hepatology (2021)
-
Identification of triptonide as a therapeutic agent for triple negative breast cancer treatment
Scientific Reports (2021)
-
LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer
Journal of Experimental & Clinical Cancer Research (2019)