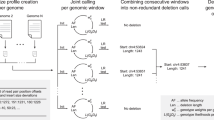
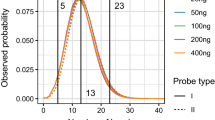
Abstract
SNP genotyping has emerged as a technology to incorporate copy number variants (CNVs) into genetic analyses of human traits. However, the extent to which SNP platforms accurately capture CNVs remains unclear. Using independent, sequence-based CNV maps, we find that commonly used SNP platforms have limited or no probe coverage for a large fraction of CNVs. Despite this, in 9 samples we inferred 368 CNVs using Illumina SNP genotyping data and experimentally validated over two-thirds of these. We also developed a method (SNP-Conditional Mixture Modeling, SCIMM) to robustly genotype deletions using as few as two SNP probes. We find that HapMap SNPs are strongly correlated with 82% of common deletions, but the newest SNP platforms effectively tag about 50%. We conclude that currently available genome-wide SNP assays can capture CNVs accurately, but improvements in array designs, particularly in duplicated sequences, are necessary to facilitate more comprehensive analyses of genomic variation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Kidd, J.M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Cooper, G.M., Nickerson, D.A. & Eichler, E.E. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 39, S22–S29 (2007).
Singleton, A.B. et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science 302, 841 (2003).
Gonzalez, E. et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307, 1434–1440 (2005).
Sharp, A.J. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 38, 1038–1042 (2006).
Perry, G.H. et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007).
Walsh, T. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320, 539–543 (2008).
Estivill, X. & Armengol, L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 3, 1787–1799 (2007).
Shaffer, L.G. & Lupski, J.R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34, 297–329 (2000).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Conrad, D.F., Andrews, T.D., Carter, N.P., Hurles, M.E. & Pritchard, J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 38, 75–81 (2006).
Locke, D.P. et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275–290 (2006).
McCarroll, S.A. et al. Common deletion polymorphisms in the human genome. Nat. Genet. 38, 86–92 (2006).
Peiffer, D.A. et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 16, 1136–1148 (2006).
Komura, D. et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 16, 1575–1584 (2006).
Colella, S. et al. QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 35, 2013–2025 (2007).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Day, N., Hemmaplardh, A., Thurman, R.E., Stamatoyannopoulos, J.A. & Noble, W.S. Unsupervised segmentation of continuous genomic data. Bioinformatics 23, 1424–1426 (2007).
Sharp, A.J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005).
She, X. et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431, 927–930 (2004).
Dempster, A.P., Laird, N.M. & Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Statist. Soc. Ser. B. Methodological 39, 1–38 (1977).
Newman, T.L. et al. High-throughput genotyping of intermediate-size structural variation. Hum. Mol. Genet. 15, 1159–1167 (2006).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Eichler, E.E. et al. Completing the map of human genetic variation. Nature 447, 161–165 (2007).
Korbel, J.O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007).
de Smith, A.J. et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum. Mol. Genet. 16, 2783–2794 (2007).
Schwarz, G. Estimating the dimension of a model. Annals of Statistics 6, 461–464 (1978).
Acknowledgements
We thank D. Peiffer and colleagues at Illumina for sharing Human 1M and HumanHap 550K genotyping data. We apologize to all colleagues whose work we could not cite because of space constraints. G.M.C. is supported by a Merck, Jane Coffin Childs Memorial Fund Postdoctoral Fellowship. T.Z. acknowledges support from the National Human Genome Research Institute (NHGRI) Interdisciplinary Training in Genomic Sciences grant T32 HG00035. J.M.K. is supported by a National Science Foundation graduate fellowship. This work was supported by the National Heart, Lung, and Blood Institute Programs for Genomic Applications grant HL066682 to D.A.N. and NHGRI grant HG004120 to E.E.E. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1, 3–6, 9, 10 and Supplementary Figures 1–4 (PDF 1667 kb)
Rights and permissions
About this article
Cite this article
Cooper, G., Zerr, T., Kidd, J. et al. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 40, 1199–1203 (2008). https://doi.org/10.1038/ng.236
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.236
This article is cited by
-
A comprehensive analysis of copy number variations in diverse apple populations
BMC Genomics (2023)
-
Assessment of linkage disequilibrium patterns between structural variants and single nucleotide polymorphisms in three commercial chicken populations
BMC Genomics (2022)
-
A map of copy number variations in the Tunisian population: a valuable tool for medical genomics in North Africa
npj Genomic Medicine (2021)
-
Benchmarking germline CNV calling tools from exome sequencing data
Scientific Reports (2021)
-
Functional and population genetic features of copy number variations in two dairy cattle populations
BMC Genomics (2020)