Abstract
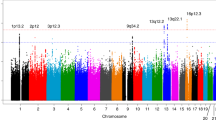
We performed a multistage genome-wide association study including 7,683 individuals with pancreatic cancer and 14,397 controls of European descent. Four new loci reached genome-wide significance: rs6971499 at 7q32.3 (LINC-PINT, per-allele odds ratio (OR) = 0.79, 95% confidence interval (CI) 0.74–0.84, P = 3.0 × 10−12), rs7190458 at 16q23.1 (BCAR1/CTRB1/CTRB2, OR = 1.46, 95% CI 1.30–1.65, P = 1.1 × 10−10), rs9581943 at 13q12.2 (PDX1, OR = 1.15, 95% CI 1.10–1.20, P = 2.4 × 10−9) and rs16986825 at 22q12.1 (ZNRF3, OR = 1.18, 95% CI 1.12–1.25, P = 1.2 × 10−8). We identified an independent signal in exon 2 of TERT at the established region 5p15.33 (rs2736098, OR = 0.80, 95% CI 0.76–0.85, P = 9.8 × 10−14). We also identified a locus at 8q24.21 (rs1561927, P = 1.3 × 10−7) that approached genome-wide significance located 455 kb telomeric of PVT1. Our study identified multiple new susceptibility alleles for pancreatic cancer that are worthy of follow-up studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 (2014).
Ferlay, J. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur. J. Cancer 49, 1374–1403 (2013).
Hidalgo, M. Pancreatic cancer. N. Engl. J. Med. 362, 1605–1617 (2010).
Klein, A.P. Genetic susceptibility to pancreatic cancer. Mol. Carcinog. 51, 14–24 (2012).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990 (2009).
Petersen, G.M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Wu, C. et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat. Genet. 44, 62–66 (2012).
Low, S.K. et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS ONE 5, e11824 (2010).
Wang, Z. et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat. Genet. 44, 6–7 (2012).
Campa, D. et al. Genetic susceptibility to pancreatic cancer and its functional characterisation: the PANcreatic Disease ReseArch (PANDoRA) consortium. Dig. Liver Dis. 45, 95–99 (2013).
Innocenti, F. et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin. Cancer Res. 18, 577–584 (2012).
Adams, J.C., Seed, B. & Lawler, J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 17, 4964–4974 (1998).
Fernandez-Zapico, M.E. et al. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem. J. 435, 529–537 (2011).
Small, K.S. et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 43, 561–564 (2011).
Barrett, A., Pellet-Many, C., Zachary, I.C., Evans, I.M. & Frankel, P. p130Cas: a key signalling node in health and disease. Cell. Signal. 25, 766–777 (2013).
Cabodi, S., del Pilar Camacho-Leal, M., Di Stefano, P. & Defilippi, P. Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer 10, 858–870 (2010).
Whitcomb, D.C. & Lowe, M.E. Human pancreatic digestive enzymes. Dig. Dis. Sci. 52, 1–17 (2007).
Whitcomb, D.C. Genetic aspects of pancreatitis. Annu. Rev. Med. 61, 413–424 (2010).
Chen, J.M. & Ferec, C. Chronic pancreatitis: genetics and pathogenesis. Annu. Rev. Genomics Hum. Genet. 10, 63–87 (2009).
Barrett, J.C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Morris, A.P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Harder, M.N. et al. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J. Clin. Endocrinol. Metab. 98, E801–E806 (2013).
't Hart, L.M. et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes 62, 3275–3281 (2013).
Stoffers, D.A., Zinkin, N.T., Stanojevic, V., Clarke, W.L. & Habener, J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15, 106–110 (1997).
MacDonald, R.J., Swift, G.H. & Real, F.X. Transcriptional control of acinar development and homeostasis. Prog. Mol. Biol. Transl. Sci. 97, 1–40 (2010).
Vaxillaire, M., Bonnefond, A. & Froguel, P. The lessons of early-onset monogenic diabetes for the understanding of diabetes pathogenesis. Best Pract. Res. Clin. Endocrinol. Metab. 26, 171–187 (2012).
Ohlsson, H., Karlsson, K. & Edlund, T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12, 4251–4259 (1993).
Manning, A.K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 (2012).
Hao, H.X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin–sensitive manner. Nature 485, 195–200 (2012).
Antoni, L., Sodha, N., Collins, I. & Garrett, M.D. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat. Rev. Cancer 7, 925–936 (2007).
Gronwald, J. et al. Cancer risks in first-degree relatives of CHEK2 mutation carriers: effects of mutation type and cancer site in proband. Br. J. Cancer 100, 1508–1512 (2009).
Fearnhead, P. SequenceLDhot: detecting recombination hotspots. Bioinformatics 22, 3061–3066 (2006).
Ballew, B.J. & Savage, S.A. Updates on the biology and management of dyskeratosis congenita and related telomere biology disorders. Expert Rev. Hematol. 6, 327–337 (2013).
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013).
James, M.A., Vikis, H.G., Tate, E., Rymaszewski, A.L. & You, M. CRR9/CLPTM1L regulates cell survival signaling and is required for Ras transformation and lung tumorigenesis. Cancer Res. 74, 1116–1127 (2014).
Jia, J. et al. CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res. 74, 2785–2795 (2014).
Rafnar, T. et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 41, 221–227 (2009).
Speedy, H.E. et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 46, 56–60 (2014).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Bojesen, S.E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 45, 371–384 (2013).
Kote-Jarai, Z. et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum. Mol. Genet. 22, 2520–2528 (2013).
Grisanzio, C. & Freedman, M.L. Chromosome 8q24–associated cancers and MYC. Genes Cancer 1, 555–559 (2010).
Huppi, K., Pitt, J.J., Wahlberg, B.M. & Caplen, N.J. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front. Genet. 3, 69 (2012).
Pharoah, P.D. et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45, 362–370 (2013).
Ahmadiyeh, N. et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA 107, 9742–9746 (2010).
Meyer, K.B. et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 7, e1002165 (2011).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Yang, T.P. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26, 2474–2476 (2010).
Westra, H.J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Lee, A.H., Heidtman, K., Hotamisligil, G.S. & Glimcher, L.H. Dual and opposing roles of the unfolded protein response regulated by IRE1α and XBP1 in proinsulin processing and insulin secretion. Proc. Natl. Acad. Sci. USA 108, 8885–8890 (2011).
Tabas-Madrid, D., Nogales-Cadenas, R. & Pascual-Montano, A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40, W478–W483 (2012).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Alavanja, M.C. et al. Cancer and noncancer risk to women in agriculture and pest control: the Agricultural Health Study. J. Occup. Med. 36, 1247–1250 (1994).
Giles, G.G. & English, D.R. The Melbourne Collaborative Cohort Study. IARC Sci. Publ. 156, 69–70 (2002).
Kolonel, L.N. et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 151, 346–357 (2000).
Lippman, S.M. et al. Designing the Selenium and Vitamin E Cancer Prevention Trial (SELECT). J. Natl. Cancer Inst. 97, 94–102 (2005).
White, E. et al. VITamins And Lifestyle cohort study: study design and characteristics of supplement users. Am. J. Epidemiol. 159, 83–93 (2004).
Porta, M. et al. In pancreatic ductal adenocarcinoma blood concentrations of some organochlorine compounds and coffee intake are independently associated with KRAS mutations. Mutagenesis 24, 513–521 (2009).
Samanic, C. et al. Smoking and bladder cancer in Spain: effects of tobacco type, timing, environmental tobacco smoke, and gender. Cancer Epidemiol. Biomarkers Prev. 15, 1348–1354 (2006).
Berndt, S.I. et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 45, 868–876 (2013).
Rothman, N. et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 42, 978–984 (2010).
Figueroa, J.D. et al. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum. Mol. Genet. 23, 1387–1398 (2014).
De Vivo, I. et al. Genome-wide association study of endometrial cancer in E2C2. Hum. Genet. 133, 211–224 (2014).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
de Bakker, P.I. et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17, R122–R128 (2008).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Wu, C. et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat. Genet. 44, 1090–1097 (2012).
Thornquist, M.D. et al. Statistical design and monitoring of the Carotene and Retinol Efficacy Trial (CARET). Control. Clin. Trials 14, 308–324 (1993).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Fearnhead, P., Harding, R.M., Schneider, J.A., Myers, S. & Donnelly, P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics 167, 2067–2081 (2004).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
Crawford, D.C. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36, 700–706 (2004).
Luna, A. & Nicodemus, K.K. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics 23, 774–776 (2007).
Lee, S.H., Wray, N.R., Goddard, M.E. & Visscher, P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011).
Acknowledgements
This project was funded in whole or in part with federal funds from the National Cancer Institute (NCI), US National Institutes of Health (NIH) under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, and mention of trade names, commercial products or organizations does not imply endorsement by the US government. Major support for PanScan III sample identification and processing was provided by the Lustgarten Foundation for Pancreatic Cancer Research. Additional support was received from NIH/NCI K07 CA140790, the American Society of Clinical Oncology Conquer Cancer Foundation, the Howard Hughes Medical Institute, the Lustgarten Foundation, the Robert T. and Judith B. Hale Fund for Pancreatic Cancer Research and Promises for Purple to B.M.W. A full list of acknowledgments for each participating study is provided in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
B.M.W., C.R., P.K., C.K., G.M.P., P.H., C.F., S.J.C., R.S.S.-S. and L.T.A. organized and designed the study. B.M.W., C.R., F.C., L.B., R.S.S.-S. and L.T.A. conducted and supervised the genotyping of samples. B.M.W., C.R., P.K., C.K., Z.W., R.B., R.S.S.-S. and L.T.A. contributed to the design and execution of statistical analyses. B.M.W., R.S.S.-S. and L.T.A. wrote the first draft of the manuscript. B.M.W., C.R., P.K., C.K., G.M.P., A.A.A., L.B.-F., P.M.B., J.B., F.C., E.J.D., S.G., G.G.G., G.E.G., P.J.G., E.J.J., A.K., A.P.K., L.N.K., M.H.K., D.L., N.M., S.H.O., H.A.R., H.D.S., K.V., E.W., W.Z., C.C.A., D.A., G.A., M.A.A., D.B., S.I.B., M.-C.B.-R., M.B., M.W.B., H.B.B.-d.-M., P.B., D.C., N.E.C., G.C., C.C., M.C., E.C., J.E., N.F., J.M.G., N.A.G., E.L.G., M. Goggins, M.J.G., M. Gross, C.A.H., M.H., K.J.H., B.E.H., E.A.H., N.H., D.J.H., F.I., M.J., R.K., T.J.K., K.-T.K., E.A.K., M.K., V.K., J.K., R.C.K., A.L., M.T.L., S.L., L.L.M., A.M., S.M., R.L.M., Y.N., A.L.O., K.O., A.V.P., P.H.M.P., U.P., R.P., A.P., M.P., F.X.R., E.R., N.R., A.S., X.-O.S., D.T.S., P.S., M.S., R.T.-W., P.R.T., G.E.T., M.T., A.T., G.S.T., D.T., P.V., J.W.-W., N.W., C.W., H.Y., K.Y., A.Z.-J., R.H., P.H., C.F., S.J.C., R.S.S.-S. and L.T.A. conducted the epidemiological studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Flow diagram of PanScan III study design
A schematic figure showing stage 1, stage 2 and replication stage with a total of 7,683 case and 14,397 control subjects included in the final analysis. Numbers of cases and controls in each stage are indicated as well as the array type, imputation and P- value threshold for SNPs moved forward to replication.
Supplementary Figure 2 Risk allele counts (a) and odds ratios (95% confidence intervals) (b) for pancreatic cancer for a genetic risk score of the ten susceptibility loci identified in PanScan I, II, and III.
(a) Percentage of cases and controls with each total number of risk alleles. Absolute number of participants provided above each vertical bar. (b) Results from unconditional logistic regression of the pancreatic cancer genotype score in participants from Stage 1 and Stage 2 of the PanScan III GWAS. For stage 1, model adjusted for age, sex, geographic region, and significant principal components. For stage 2, model adjusted for age, sex, study, and significant principal components. Referent is the most prevalent risk allele count in controls (n=10 risk alleles).
Supplementary Figure 3 Plot of estimated admixture for individuals in stage 1 of PanScan III GWAS.
For details, see Online Methods. Individuals with <80% European ancestry were excluded from the main association analysis. Individuals with Asian ancestry from SMWHS were analyzed separately and included case and control subjects from SMWHS in stages 1 and 2 of PanScan III and control subjects from SMWHS that were previously genotyped.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–12 and Supplementary Note (PDF 2802 kb)
Rights and permissions
About this article
Cite this article
Wolpin, B., Rizzato, C., Kraft, P. et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 46, 994–1000 (2014). https://doi.org/10.1038/ng.3052
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3052
This article is cited by
-
Genetic and other risk factors for pancreatic ductal adenocarcinoma (PDAC)
Familial Cancer (2024)
-
Association of genetic risk and lifestyle with pancreatic cancer and their age dependency: a large prospective cohort study in the UK Biobank
BMC Medicine (2023)
-
A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories
Nature Medicine (2023)
-
Exploring the Neandertal legacy of pancreatic ductal adenocarcinoma risk in Eurasians
Biological Research (2023)
-
Current status of inherited pancreatic cancer
Hereditary Cancer in Clinical Practice (2022)