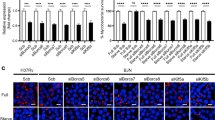
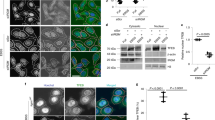
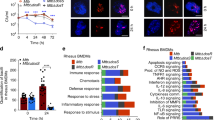
Abstract
Autophagy is emerging as a crucial defense mechanism against bacteria, but the host intracellular sensors responsible for inducing autophagy in response to bacterial infection remain unknown. Here we demonstrated that the intracellular sensors Nod1 and Nod2 are critical for the autophagic response to invasive bacteria. By a mechanism independent of the adaptor RIP2 and transcription factor NF-κB, Nod1 and Nod2 recruited the autophagy protein ATG16L1 to the plasma membrane at the bacterial entry site. In cells homozygous for the Crohn's disease–associated NOD2 frameshift mutation, mutant Nod2 failed to recruit ATG16L1 to the plasma membrane and wrapping of invading bacteria by autophagosomes was impaired. Our results link bacterial sensing by Nod proteins to the induction of autophagy and provide a functional link between Nod2 and ATG16L1, which are encoded by two of the most important genes associated with Crohn's disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mizushima, N., Levine, B., Cuervo, A.M. & Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Reggiori, F. & Klionsky, D.J. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17, 415–422 (2005).
Hussey, S., Travassos, L.H. & Jones, N.L. Autophagy as an emerging dimension to adaptive and innate immunity. Semin. Immunol. 21, 233–241 (2009).
Marx, J. Autophagy: is it cancer's friend or foe? Science 312, 1160–1161 (2006).
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Birmingham, C.L. et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy 3, 442–451 (2007).
Ogawa, M. et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005).
Gutierrez, M.G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Birmingham, C.L. & Brumell, J.H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy 2, 156–158 (2006).
Carneiro, L., Magalhaes, J., Tattoli, I., Philpott, D. & Travassos, L. Nod-like proteins in inflammation and disease. J. Pathol. 214, 136–148 (2008).
Martinon, F. & Tschopp, J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 26, 447–454 (2005).
Meylan, E., Tschopp, J. & Karin, M. Intracellular pattern recognition receptors in the host response. Nature 442, 39–44 (2006).
Chamaillard, M. et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707 (2003).
Girardin, S.E. et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587 (2003).
Girardin, S.E. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 (2003).
Girardin, S.E. et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708 (2003).
Inohara, N. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 (2003).
Inohara, N. et al. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 275, 27823–27831 (2000).
Abbott, D.W., Wilkins, A., Asara, J.M. & Cantley, L.C. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14, 2217–2227 (2004).
Hugot, J.P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Rioux, J.D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Fritz, J.H. et al. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity 26, 445–459 (2007).
Magalhaes, J.G. et al. Nod2-dependent Th2 polarization of antigen-specific immunity. J. Immunol. 181, 7925–7935 (2008).
Delgado, M.A., Elmaoued, R.A., Davis, A.S., Kyei, G. & Deretic, V. Toll-like receptors control autophagy. EMBO J. 27, 1110–1121 (2008).
Klionsky, D.J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008).
Mizushima, N., Ohsumi, Y. & Yoshimori, T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27, 421–429 (2002).
Magalhaes, J.G. et al. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 6, 1201–1207 (2005).
Opitz, B. et al. Listeria monocytogenes activated p38 MAPK and induced IL-8 secretion in a nucleotide-binding oligomerization domain 1-dependent manner in endothelial cells. J. Immunol. 176, 484–490 (2006).
Travassos, L.H. et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J. Biol. Chem. 280, 36714–36718 (2005).
Viala, J. et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5, 1166–1174 (2004).
Carneiro, L.A. et al. Shigella induces mitochondrial dysfunction and cell death in nonmyleoid cells. Cell Host Microbe 5, 123–136 (2009).
Opitz, B. et al. Nod1-mediated endothelial cell activation by Chlamydophila pneumoniae. Circ. Res. 96, 319–326 (2005).
Kim, S., La Motte-Mohs, R.N., Rudolph, D., Zuniga-Pflucker, J.C. & Mak, T.W. The role of nuclear factor-κB essential modulator (NEMO) in B cell development and survival. Proc. Natl. Acad. Sci. USA 100, 1203–1208 (2003).
Barnich, N., Aguirre, J.E., Reinecker, H.C., Xavier, R. & Podolsky, D.K. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-κB activation in muramyl dipeptide recognition. J. Cell Biol. 170, 21–26 (2005).
Lecine, P. et al. The NOD2-RICK complex signals from the plasma membrane. J. Biol. Chem. 282, 15197–15207 (2007).
Till, A. et al. A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. J. Cell Sci. 121, 487–495 (2008).
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008).
Kuballa, P., Huett, A., Rioux, J.D., Daly, M.J. & Xavier, R.J. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE 3, e3391 (2008).
Xu, Y. et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27, 135–144 (2007).
Kaneko, T. et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723 (2006).
Zaidman-Remy, A. et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473 (2006).
Yano, T. et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916 (2008).
Hofius, D. et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783 (2009).
Allaoui, A., Mounier, J., Prevost, M.C., Sansonetti, P.J. & Parsot, C. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol. Microbiol. 6, 1605–1616 (1992).
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 104, 14050–14055 (2007).
Acknowledgements
We thank K. Croitoru (University of Toronto) for Mode-K cells; N. Mizushima (Tokyo Medical and Dental Center) for ATG5-deficient MEFs; T. Mak (University of Toronto) for NEMO-deficient MEFs; R. Flavell (Yale University) for pEasyFlox; N. Mizushima (Tokyo Medical and Dental University Graduate School) for Flag–ΔN85-ATG16L1, Flag–ΔN85-ATG16L1*300A and HA-ATG5; R. Xavier (Harvard Medical School) for FL-ATG16L1–Myc and FL-ATG16L1*300A–Myc; M. D'Amato (Karolinska Institutet) for HA-Nod1-HA, HA-Nod2, HA–Nod2 L1007fs; F. Takeshita (Yokohama City University School of Medicine) for GFP-HA and pHR-SIN-CSGWΔNotI-GFP-LC3 GFP-Flag; C. Münz (University of Zurich) for rabbit polyclonal antibody to LC3 (anti-LC3 and for pHR-SIN-CSGWΔNotI-GFP-LC3); T.A. Kufer and E. Kremmer (University of Cologne and University of Munich) for rat monoclonal anti-Nod2; P. Sansonetti (Institute Pasteur) for S. flexneri M90T; D. Portnoy (University of California) for L. monocytogenes 10403S and the listeriolysin O deletion mutant; all other donors for reagents, antibodies, plasmids and bacterial strains; and M. Silverberg and J. Brumell for discussions. Supported by the Canadian Institutes for Health Research (L.H.T. and L.A.M.C.; MOP480142 to D.J.P.; and MOP81360 to S.E.G.), the Canadian Association Gastroenterology (S.H.), the University of Michigan (Y.-G.K.), Fondation Bettencourt-Schueller (J.G.M.), Fundação para Ciência e Tecnologia de Portugal (J.G.M.), the National Institutes of Health (DK61707 to G.N.), the Crohn's & Colitis Foundation of Canada (N.L.J., D.J.P. and S.E.G.) European Research Council (202283-PGN from SHAPE to VIR to I.G.B.) and the Howard Hughes Medical Institute (D.J.P.).
Author information
Authors and Affiliations
Contributions
L.H.T. and L.A.M.C. designed and did experiments, analyzed data and wrote the paper; M.R., S.H., J.G.M., L.Y., F.S., E.C., L.L.B., I.G.B., A.A. and N.L.J. did experiments; Y.-G.K. and G.N. contributed tools; and S.E.G. and D.J.P. designed experiment, analyzed data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 1076 kb)
Rights and permissions
About this article
Cite this article
Travassos, L., Carneiro, L., Ramjeet, M. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11, 55–62 (2010). https://doi.org/10.1038/ni.1823
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1823
This article is cited by
-
The NLR gene family: from discovery to present day
Nature Reviews Immunology (2023)
-
NOD2 inhibits the proliferation of esophageal adenocarcinoma cells through autophagy
Journal of Cancer Research and Clinical Oncology (2023)
-
Autophagic reprogramming of bone marrow–derived macrophages
Immunologic Research (2023)
-
Therapeutic potential of autophagy in immunity and inflammation: current and future perspectives
Pharmacological Reports (2023)
-
Polymorphisms in autophagy genes are genetic susceptibility factors in glioblastoma development
BMC Cancer (2022)