Abstract
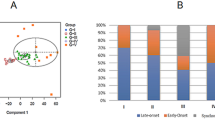
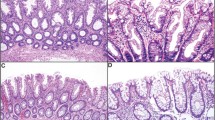
Colon cancer is a clinically diverse disease. This heterogeneity makes it difficult to determine which patients will benefit most from adjuvant therapy and impedes the development of new targeted agents1. More insight into the biological diversity of colon cancers, especially in relation to clinical features, is therefore needed. We demonstrate, using an unsupervised classification strategy involving over 1,100 individuals with colon cancer, that three main molecularly distinct subtypes can be recognized. Two subtypes have been previously identified and are well characterized (chromosomal-instable and microsatellite-instable cancers)2. The third subtype is largely microsatellite stable and contains relatively more CpG island methylator phenotype–positive carcinomas but cannot be identified on the basis of characteristic mutations. We provide evidence that this subtype relates to sessile-serrated adenomas, which show highly similar gene expression profiles, including upregulation of genes involved in matrix remodeling and epithelial-mesenchymal transition. The identification of this subtype is crucial, as it has a very unfavorable prognosis and, moreover, is refractory to epidermal growth factor receptor–targeted therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wolpin, B.M. & Mayer, R.J. Systemic treatment of colorectal cancer. Gastroenterology 134, 1296–1310 (2008).
Markowitz, S.D. & Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 361, 2449–2460 (2009).
Verhaak, R.G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Julien, S. et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin. Cancer Res. 18, 5314–5328 (2012).
Uronis, J.M. et al. Histological and molecular evaluation of patient-derived colorectal cancer explants. PLoS ONE 7, e38422 (2012).
Calon, A. et al. Dependency of colorectal cancer on a TGF-β–driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012).
Pagès, F. et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 27, 5944–5951 (2009).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Shen, L. et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc. Natl. Acad. Sci. USA 104, 18654–18659 (2007).
Schlicker, A. et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med. Genomics 5, 66 (2012).
Sugai, T. et al. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J. Mol. Diagn. 8, 193–201 (2006).
Lascorz, J., Chen, B., Hemminki, K. & Forsti, A. Consensus pathways implicated in prognosis of colorectal cancer identified through systematic enrichment analysis of gene expression profiling studies. PLoS ONE 6, e18867 (2011).
Salazar, R. et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J. Clin. Oncol. 29, 17–24 (2011).
Popovici, V. et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J. Clin. Oncol. 30, 1288–1295 (2012).
Gray, R.G. et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J. Clin. Oncol. 29, 4611–4619 (2011).
Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567–6572 (2002).
Khambata-Ford, S. et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J. Clin. Oncol. 25, 3230–3237 (2007).
Boparai, K.S. et al. A serrated colorectal cancer pathway predominates over the classic WNT pathway in patients with hyperplastic polyposis syndrome. Am. J. Pathol. 178, 2700–2707 (2011).
Kambara, T. et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53, 1137–1144 (2004).
Van der Flier, L.G. et al. The intestinal Wnt/TCF signature. Gastroenterology 132, 628–632 (2007).
Taube, J.H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA 107, 15449–15454 (2010).
Curran, S. et al. Matrix metalloproteinase/tissue inhibitors of matrix metalloproteinase phenotype identifies poor prognosis colorectal cancers. Clin. Cancer Res. 10, 8229–8234 (2004).
Shioiri, M. et al. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br. J. Cancer 94, 1816–1822 (2006).
Spaderna, S. et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 131, 830–840 (2006).
de Sousa E Melo, F. et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9, 476–485 (2011).
Bruna, A. et al. TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat. Commun. 3, 1055 (2012).
Bennecke, M. et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell 18, 135–146 (2010).
Leystra, A.A. et al. Mice expressing activated PI3K rapidly develop advanced colon cancer. Cancer Res. 72, 2931–2936 (2012).
Merlos-Suárez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011).
Jorissen, R.N. et al. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin. Cancer Res. 14, 8061–8069 (2008).
Tsuji, S. et al. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br. J. Cancer 106, 126–132 (2012).
Wagner, K.W. et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 13, 1070–1077 (2007).
McCall, M.N., Bolstad, B.M. & Irizarry, R.A. Frozen robust multiarray analysis (fRMA). Biostatistics 11, 242–253 (2010).
McCall, M.N., Uppal, K., Jaffee, H.A., Zilliox, M.J. & Irizarry, R.A. The Gene Expression Barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res. 39, D1011–D1015 (2011).
Monti, S., Tamayo, P., Mesirov, J. & Golub, T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 52, 91–118 (2003).
Tibshirani, R., Walther, G. & Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Series B Stat. Methodol. 63, 411–423 (2001).
Rousseeuw, P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567–6572 (2002).
Johnson, W.E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Clark-Langone, K.M., Sangli, C., Krishnakumar, J. & Watson, D. Translating tumor biology into personalized treatment planning: analytical performance characteristics of the Oncotype DX Colon Cancer Assay. BMC Cancer 10, 691 (2010).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Ruifrok, A.C. & Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23, 291–299 (2001).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Weisenberger, D.J. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38, 787–793 (2006).
Acknowledgements
We wish to thank P. van Sluijs and R. Volckmann for assistance with deriving gene expression profiles, J. Lascorz for sharing his collection of prognostic signatures, S. Kozar for important comments on the manuscript, and K. Punt, I. Nagtegaal and P. Kuppen for valuable insights from their patient cohorts. This work was supported by a Vici grant from the Netherlands Organisation for Scientific Research to J.P.M. and Dutch Cancer Society grants (2009-4416 and 2012-5735). L.V. is supported by a Koningin Wilhelmina Fonds (KWF, Dutch Cancer Society Fellowship).
Author information
Authors and Affiliations
Contributions
F.D.S.E.M., M.J., E.F., A.T., L.P.M.H.d.R., J.H.d.J., O.J.d.B., R.v.L., M.F.B., H.R. and M.v.d.H. performed experiments; M.J., C.J.M.v.N., J.B.T. and E.D. provided tissue and pathological assistance; F.D.S.E.M., X.W., F.M., J.P.M. and L.V. analyzed the data; and J.P.M. and L.V. supervised the project and wrote the manuscript. All authors approved the content of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 3439 kb)
Supplementary Tables
Supplementary Tables 1–10 (XLSX 231 kb)
Rights and permissions
About this article
Cite this article
De Sousa E Melo, F., Wang, X., Jansen, M. et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 19, 614–618 (2013). https://doi.org/10.1038/nm.3174
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3174
This article is cited by
-
Towards precision oncology discovery: four less known genes and their unknown interactions as highest-performed biomarkers for colorectal cancer
npj Precision Oncology (2024)
-
KRAS allelic imbalance drives tumour initiation yet suppresses metastasis in colorectal cancer in vivo
Nature Communications (2024)
-
Molecular pathological classification of colorectal cancer—an update
Virchows Archiv (2024)
-
ZEB1 hypermethylation is associated with better prognosis in patients with colon cancer
Clinical Epigenetics (2023)
-
A comparative study of clustering methods on gene expression data for lung cancer prognosis
BMC Research Notes (2023)