Abstract
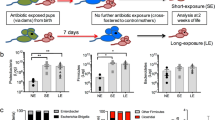
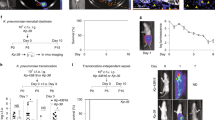
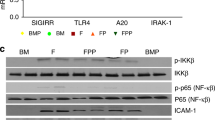
Neonatal colonization by microbes, which begins immediately after birth, is influenced by gestational age and the mother's microbiota and is modified by exposure to antibiotics1. In neonates, prolonged duration of antibiotic therapy is associated with increased risk of late-onset sepsis (LOS)2, a disorder controlled by neutrophils3. A role for the microbiota in regulating neutrophil development and susceptibility to sepsis in the neonate remains unclear. We exposed pregnant mouse dams to antibiotics in drinking water to limit transfer of maternal microbes to the neonates. Antibiotic exposure of dams decreased the total number and composition of microbes in the intestine of the neonates. This was associated with decreased numbers of circulating and bone marrow neutrophils and granulocyte/macrophage–restricted progenitor cells in the bone marrow of antibiotic-treated and germ-free neonates. Antibiotic exposure of dams reduced the number of interleukin-17 (IL-17)-producing cells in the intestine and production of granulocyte colony–stimulating factor (G-CSF). Granulocytopenia was associated with impaired host defense and increased susceptibility to Escherichia coli K1 and Klebsiella pneumoniae sepsis in antibiotic-treated neonates, which could be partially reversed by administration of G-CSF. Transfer of a normal microbiota into antibiotic-treated neonates induced IL-17 production by group 3 innate lymphoid cells (ILCs) in the intestine, increasing plasma G-CSF levels and neutrophil numbers in a Toll-like receptor 4 (TLR4)– and myeloid differentiation factor 88 (MyD88)–dependent manner and restored IL-17–dependent resistance to sepsis. Specific depletion of ILCs prevented IL-17– and G-CSF–dependent granulocytosis and resistance to sepsis. These data support a role for the intestinal microbiota in regulation of granulocytosis, neutrophil homeostasis and host resistance to sepsis in neonates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Penders, J. et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 (2006).
Kuppala, V.S., Meinzen-Derr, J., Morrow, A.L. & Schibler, K.R. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J. Pediatr. 159, 720–725 (2011).
Sarkar, S., Bhagat, I., Hieber, S. & Donn, S.M. Can neutrophil responses in very low birth weight infants predict the organisms responsible for late-onset bacterial or fungal sepsis? J. Perinatol. 26, 501–505 (2006).
Schwiertz, A. et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr. Res. 54, 393–399 (2003).
Pflughoeft, K.J. & Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 7, 99–122 (2012).
Mai, V. et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 8, e52876 (2013).
Palmer, C., Bik, E.M., DiGiulio, D.B., Relman, D.A. & Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Vael, C. & Desager, K. The importance of the development of the intestinal microbiota in infancy. Curr. Opin. Pediatr. 21, 794–800 (2009).
Koenig, J.E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 108 (suppl. 1), 4578–4585 (2011).
Alm, B. et al. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics 121, 697–702 (2008).
Clark, R.H., Bloom, B.T., Spitzer, A.R. & Gerstmann, D.R. Empiric use of ampicillin and cefotaxime, compared with ampicillin and gentamicin, for neonates at risk for sepsis is associated with an increased risk of neonatal death. Pediatrics 117, 67–74 (2006).
Manroe, B.L., Weinberg, A.G., Rosenfeld, C.R. & Browne, R. The neonatal blood count in health and disease. I. Reference values for neutrophilic cells. J. Pediatr. 95, 89–98 (1979).
Gessler, P. et al. Neonatal neutropenia in low birthweight premature infants. Am. J. Perinatol. 12, 34–38 (1995).
Martin, C. et al. Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity 19, 583–593 (2003).
Pluschke, G. & Pelkonen, S. Host factors in the resistance of newborn mice to K1 Escherichia coli infection. Microb. Pathog. 4, 93–102 (1988).
Cohen-Wolkowiez, M. et al. Early and late onset sepsis in late preterm infants. Pediatr. Infect. Dis. J. 28, 1052–1056 (2009).
Lieschke, G.J. et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 (1994).
Sekirov, I. et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008).
Clarke, T.B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Schwarzenberger, P. et al. Requirement of endogenous stem cell factor and granulocyte-colony-stimulating factor for IL-17–mediated granulopoiesis. J. Immunol. 164, 4783–4789 (2000).
Ye, P. et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194, 519–527 (2001).
Ivanov, I.I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Ivanov, I.I. et al. Specific microbiota direct the differentiation of IL-17–producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Denning, T.L., Wang, Y.C., Patel, S.R., Williams, I.R. & Pulendran, B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17–producing T cell responses. Nat. Immunol. 8, 1086–1094 (2007).
Mombaerts, P. et al. RAG-1–deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Spits, H. et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013).
Sawa, S. et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science 330, 665–669 (2010).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23–dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Medzhitov, R. TLR-mediated innate immune recognition. Semin. Immunol. 19, 1–2 (2007).
Schnare, M. et al. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2, 947–950 (2001).
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359 (2011).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Yamamoto, M. et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Bugl, S. et al. Steady-state neutrophil homeostasis is dependent on TLR4/TRIF signaling. Blood 121, 723–733 (2013).
Elinav, E. et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 13, 759–771 (2013).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Mazmanian, S.K., Liu, C.H., Tzianabos, A.O. & Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Lotz, M. et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984 (2006).
Daenen, S., Goris, H., de Boer, F., Halie, M.R. & van der Waaij, D. Recovery of murine myelopoiesis after cytostatic reduction by Ara-C. Effect of bacitracin-induced changes in the intestinal microflora and influence of timing. Leuk. Res. 15, 1013–1018 (1991).
Hill, D.A. et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158 (2010).
Rudi, K., Tannaes, T. & Vatn, M. Temporal and spatial diversity of the tap water microbiota in a Norwegian hospital. Appl. Environ. Microbiol. 75, 7855–7857 (2009).
Oliwa-Stasiak, K., Kolaj-Robin, O. & Adley, C.C. Development of real-time PCR assays for detection and quantification of Bacillus cereus group species: differentiation of B. weihenstephanensis and rhizoid B. pseudomycoides isolates from milk. Appl. Environ. Microbiol. 77, 80–88 (2011).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Mei, J. et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest. 122, 974–986 (2012).
Acknowledgements
We thank D. Artis (Department of Microbiology and Pathology, University of Pennsylvania) for providing GF mice. We thank N. Butz for her assistance with microbial DNA isolation. We thank the Children's Hospital of Philadelphia Research Institute Flow Cytometry and Cell Sorting Core Laboratory for technical advice and support. We thank S. Guttentag, K. Hudock and C. Hergott for their helpful comments. H.S.D. is supported by 5T32HD060556, P.M.O. is supported by 5R01AI093566, J.K.K. is supported by 5R01HL062052, 3R37HL079142 and 5P60AA009803, J.N.W. is supported by 1R01AI105168 and 5R01AI038446 and G.S.W. is supported by 1R01AI099479 and 5R01HL105834.
Author information
Authors and Affiliations
Contributions
H.S.D., J.N.W. and G.S.W. conceived of the study. H.S.D. and G.S.W. designed the experiments. P.M.O. and J.K.K. provided reagents. H.S.D., O.R.M., Y.L., N.D., J.M. and C.E.O. carried out experiments. H.S.D. and G.S.W. analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 1248 kb)
Rights and permissions
About this article
Cite this article
Deshmukh, H., Liu, Y., Menkiti, O. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 20, 524–530 (2014). https://doi.org/10.1038/nm.3542
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3542
This article is cited by
-
Temporal dynamics of the fecal microbiome in female pigs from early life through estrus, parturition, and weaning of the first litter of piglets
Animal Microbiome (2024)
-
Prenylcysteine oxidase 1 like protein is required for neutrophil bactericidal activities
Nature Communications (2023)
-
Early gut microbiological changes and metabolomic changes in patients with sepsis: a preliminary study
International Microbiology (2023)
-
Mechanistic Insights into Immune-Microbiota Interactions and Preventive Role of Probiotics Against Autoimmune Diabetes Mellitus
Probiotics and Antimicrobial Proteins (2023)
-
Less is more: Antibiotics at the beginning of life
Nature Communications (2023)