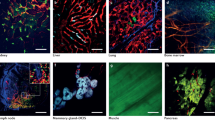
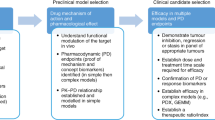
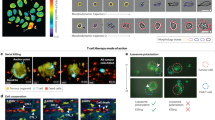
Key Points
-
Advanced fluorescence-based imaging, three-dimensional intermediate systems and intravital mouse models can be integrated into the standard drug project operating model (DPOM) to better inform the development and selection of new candidate drugs.
-
Intermediate systems provide initial three-dimensional imaging early in the drug discovery process to support translational cancer research in more physiologically relevant in vitro settings and identify deficient or ineffective drug strategies earlier in the drug discovery pipeline.
-
In vivo advanced imaging techniques can be used to assess more complex questions, such as transient protein–protein interactions or molecular, cell or tissue-specific dynamics in response to drug treatment in live tissue.
-
Biosensors are now providing dynamic and reversible fluorescence-based readouts of drug targeting, allowing drug turnover, clearance and dissociation to be monitored in real-time.
-
Stromal targeting of the tumour microenvironment is a vital aspect of cancer drug development, which can be quantified using advanced imaging techniques, such as second harmonic generation (SHG), third harmonic generation (THG) and fluorescence lifetime imaging microscopy (FLIM).
-
Longitudinal imaging through intravital imaging windows can give quantitative functional information from repeated, non-invasive imaging and drug endpoint analysis in real-time.
Abstract
Integrating biological imaging into early stages of the drug discovery process can provide invaluable readouts of drug activity within complex disease settings, such as cancer. Iterating this approach from initial lead compound identification in vitro to proof-of-principle in vivo analysis represents a key challenge in the drug discovery field. By embracing more complex and informative models in drug discovery, imaging can improve the fidelity and statistical robustness of preclinical cancer studies. In this Review, we highlight how combining advanced imaging with three-dimensional systems and intravital mouse models can provide more informative and disease-relevant platforms for cancer drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Swinney, D. C. The contribution of mechanistic understanding to phenotypic screening for first-in-class medicines. J. Biomol. Screen 18, 1186–1192 (2013).
Carragher, N. O. et al. Live cell in vitro and in vivo imaging applications: accelerating drug discovery. Pharmaceutics 3, 141–170 (2011).
Timpson, P., McGhee, E. J. & Anderson, K. I. Imaging molecular dynamics in vivo—from cell biology to animal models. J. Cell Sci. 124, 2877–2890 (2011).
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nature Rev. Drug Discov. 9, 203–214 (2010).
Tentler, J. J. et al. Patient-derived tumour xenografts as models for oncology drug development. Nature Rev. Clin. Oncol. 9, 338–350 (2012).
Kamb, A. What's wrong with our cancer models? Nature Rev. Drug Discov. 4, 161–165 (2005).
Weissleder, R. & Pittet, M. J. Imaging in the era of molecular oncology. Nature 452, 580–589 (2008).
Beerling, E., Ritsma, L., Vrisekoop, N., Derksen, P. W. & van Rheenen, J. Intravital microscopy: new insights into metastasis of tumors. J. Cell Sci. 124, 299–310 (2011).
Lee, J. A. & Berg, E. L. Neoclassic drug discovery: the case for lead generation using phenotypic and functional approaches. J. Biomol. Screen 18, 1143–1155 (2013).
Swinney, D. C. & Anthony, J. How were new medicines discovered? Nature Rev. Drug Discov. 10, 507–519 (2011). This paper reviews the contributions of target- and phenotypic-directed drug discovery in a retrospective analysis of all drugs approved by the FDA between 1999 and 2008.
Bakal, C., Aach, J., Church, G. & Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316, 1753–1756 (2007).
Vindin, H., Bischof, L., Gunning, P. & Stehn, J. Validation of an algorithm to quantify changes in actin cytoskeletal organization. J. Biomol. Screen 19, 354–368 (2013).
Cappella, P. & Gasparri, F. Highly multiplexed phenotypic imaging for cell proliferation studies. J. Biomol. Screen 19, 145–157 (2013).
Yarrow, J. C., Totsukawa, G., Charras, G. T. & Mitchison, T. J. Screening for cell migration inhibitors via automated microscopy reveals a Rho-kinase inhibitor. Chem. Biol. 12, 385–395 (2005).
Xu, G. W. et al. A high-content chemical screen identifies ellipticine as a modulator of p53 nuclear localization. Apoptosis 13, 413–422 (2008).
Young, D. W. et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nature Chem. Biol. 4, 59–68 (2008).
Rose, R. H., Briddon, S. J. & Holliday, N. D. Bimolecular fluorescence complementation: lighting up seven transmembrane domain receptor signalling networks. Br. J. Pharmacol. 159, 738–750 (2010).
Dai, J. P. et al. Drug screening for autophagy inhibitors based on the dissociation of Beclin1-Bcl2 complex using BiFC technique and mechanism of eugenol on anti-influenza A virus activity. PLoS ONE 8, e61026 (2013).
Shinjo, S., Tashiro, E. & Imoto, M. Establishment of a new detection system for the dimerization of IRE1α by BiFC assay. Biosci. Biotechnol. Biochem. 77, 1333–1336 (2013).
Filonov, G. S. & Verkhusha, V. V. A near-infrared bifc reporter for in vivo imaging of protein-protein interactions. Chem. Biol. 20, 1078–1086 (2013). This paper describes the design of the first near-infrared BiFC reporter for in vivo protein interaction studies.
Fang, D. & Kerppola, T. K. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl Acad. Sci. USA 101, 14782–14787 (2004).
Li, R. et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 73, 5742–5753 (2013).
Day, C. A., Kraft, L. J., Kang, M. & Kenworthy, A. K. Analysis of protein and lipid dynamics using confocal fluorescence recovery after photobleaching (FRAP). Curr. Protoc. Cytom. 62, 2.19.1–2.19.29 (2012).
Canel, M., Serrels, A., Anderson, K. I., Frame, M. C. & Brunton, V. G. Use of photoactivation and photobleaching to monitor the dynamic regulation of E-cadherin at the plasma membrane. Cell Adh. Migr. 4, 491–501 (2010).
Canel, M. et al. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res. 70, 9413–9422 (2010). This paper describes the combined application of intravital imaging windows with three distinct subcellular advanced techniques (photoactivation, photoswitching and FRAP) to examine tumour cell–cell adhesion strength and response to anti-invasive receptor tyrosine kinase (RTK) or endocytic drug treatment.
Serrels, A. et al. Real-time study of E-cadherin and membrane dynamics in living animals: implications for disease modeling and drug development. Cancer Res. 69, 2714–2719 (2009). This paper describes the first use of live in vivo FRAP to measure cell–cell junction dynamics in living solid tumour tissue: FRAP was used as a surrogate marker of the tumour dissociation response to therapeutic intervention.
Yamada, S., Pokutta, S., Drees, F., Weis, W. I. & Nelson, W. J. Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 (2005).
de Beco, S., Gueudry, C., Amblard, F. & Coscoy, S. Endocytosis is required for E-cadherin redistribution at mature adherens junctions. Proc. Natl Acad. Sci. USA 106, 7010–7015 (2009).
Cavey, M., Rauzi, M., Lenne, P. F. & Lecuit, T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature 453, 751–756 (2008).
Daddysman, M. K. & Fecko, C. J. Revisiting point FRAP to quantitatively characterize anomalous diffusion in live cells. J. Phys. Chem. B 117, 1241–1251 (2013).
Dieteren, C. E. et al. Solute diffusion is hindered in the mitochondrial matrix. Proc. Natl Acad. Sci. USA 108, 8657–8662 (2011).
Andrews, P. D. et al. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253–268 (2004).
Famulski, J. K. & Chan, G. K. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr. Biol. 17, 2143–2149 (2007).
Khodjakov, A. & Rieder, C. L. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146, 585–596 (1999).
Vink, M. et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16, 755–766 (2006).
Mueller, F., Wach, P. & McNally, J. G. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 94, 3323–3339 (2008).
Mazza, D., Abernathy, A., Golob, N., Morisaki, T. & McNally, J. G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 (2012).
Misteli, T., Gunjan, A., Hock, R., Bustin, M. & Brown, D. T. Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 (2000).
Kang, M., Day, C. A., DiBenedetto, E. & Kenworthy, A. K. A quantitative approach to analyze binding diffusion kinetics by confocal FRAP. Biophys. J. 99, 2737–2747 (2010).
Nouar, R., Devred, F., Breuzard, G. & Peyrot, V. FRET and FRAP imaging: approaches to characterise tau and stathmin interactions with microtubules in cells. Biol. Cell 105, 149–161 (2013).
Murthy, K. & Wadsworth, P. Dual role for microtubules in regulating cortical contractility during cytokinesis. J. Cell Sci. 121, 2350–2359 (2008).
Kraft, L. J. & Kenworthy, A. K. Imaging protein complex formation in the autophagy pathway: analysis of the interaction of LC3 and Atg4B(C74A) in live cells using Forster resonance energy transfer and fluorescence recovery after photobleaching. J. Biomed. Opt. 17, 011008 (2012).
Schneider, K. et al. Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Res. 41, 4860–4876 (2013).
Schermelleh, L. et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35, 4301–4312 (2007).
Spada, F. et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176, 565–571 (2007).
Agasti, S. S. et al. Dual imaging and photoactivated nanoprobe for controlled cell tracking. Small 9, 222–227 (2013).
Wang, X., He, L., Wu, Y. I., Hahn, K. M. & Montell, D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature Cell Biol. 12, 591–597 (2010).
Yoo, S. K. et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236 (2010).
Frost, N. A., Lu, H. E. & Blanpied, T. A. Optimization of cell morphology measurement via single-molecule tracking PALM. PLoS ONE 7, e36751 (2012).
Roy, S., Yang, G., Tang, Y. & Scott, D. A. A simple photoactivation and image analysis module for visualizing and analyzing axonal transport with high temporal resolution. Nature Protoc. 7, 62–68 (2012).
Caswell, P. T. et al. Rab25 associates with α5β1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 13, 496–510 (2007).
Amornphimoltham, P. et al. Rab25 regulates invasion and metastasis in head and neck cancer. Clin. Cancer Res. 19, 1375–1388 (2013).
Ritsma, L. et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci. Transl Med. 4, 158ra145 (2012). This paper provides an insight into the progressive nature and capacity of intravital imaging windows to monitor late stages of metastasis from deep within the body cavity at high resolution, revealing a time-dependent aspect to when anti-migratory targeting can be effective.
Kedrin, D. et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nature Methods 5, 1019–1021 (2008). This paper describes the combined application of intravital imaging windows with photoactivation for repeated imaging and tracking of tumour population dynamics in mammary tumours.
Yu, X. et al. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol. 199, 527–544 (2012).
Deakin, N. O., Ballestrem, C. & Turner, C. E. Paxillin and Hic-5 interaction with vinculin is differentially regulated by Rac1 and RhoA. PLoS ONE 7, e37990 (2012). This study demonstrates the application of FRET biosensors to provide novel insight into protein–protein interactions within cell adhesions and their distinction between 2D and 3D in vitro models.
Wouters, F. S., Verveer, P. J. & Bastiaens, P. I. Imaging biochemistry inside cells. Trends Cell Biol. 11, 203–211 (2001).
Fruhwirth, G. O. et al. How Forster resonance energy transfer imaging improves the understanding of protein interaction networks in cancer biology. Chemphyschem 12, 442–461 (2011). This is a comprehensive overview of the potential use of FRET-based biosensor imaging in cancer.
Seong, J. et al. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nature Commun. 2, 406 (2011).
Wang, Y. et al. Visualizing the mechanical activation of Src. Nature 434, 1040–1045 (2005).
Hirata, E. et al. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 125, 858–868 (2012).
Ouyang, M. et al. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J. Biol. Chem. 283, 17740–17748 (2008).
Lu, S. et al. Quantitative FRET imaging to visualize the invasiveness of live breast cancer cells. PLoS ONE 8, e58569 (2013).
Ouyang, M. et al. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res. 70, 2204–2212 (2010). This paper describes dual FLIM–FRET imaging of spectrally distinct biosensors for MT1MMP and SRC activity.
Luo, K. Q., Yu, V. C., Pu, Y. & Chang, D. C. Application of the fluorescence resonance energy transfer method for studying the dynamics of caspase-3 activation during UV-induced apoptosis in living HeLa cells. Biochem. Biophys. Res. Commun. 283, 1054–1060 (2001).
Yoshizaki, H. et al. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162, 223–232 (2003).
Gavet, O. & Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189, 247–259 (2010).
Gavet, O. & Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 18, 533–543 (2010).
Nobis, M. et al. Intravital FLIM-FRET imaging reveals dasatinib-induced spatial control of Src in pancreatic cancer. Cancer Res. 75, 4674–4686 (2013). This paper highlights the usefulness of FRET-biosensor expression in target tissue, providing a reversible and dynamic readout of target inactivation and clearance response to drug treatment in live tumours.
Milligan, G. Applications of bioluminescence- and fluorescence resonance energy transfer to drug discovery at G protein-coupled receptors. Eur. J. Pharm. Sci. 21, 397–405 (2004).
Tian, H., Ip, L., Luo, H., Chang, D. C. & Luo, K. Q. A high throughput drug screen based on fluorescence resonance energy transfer (FRET) for anticancer activity of compounds from herbal medicine. Br. J. Pharmacol. 150, 321–334 (2007).
Stockholm, D. et al. Imaging calpain protease activity by multiphoton FRET in living mice. J. Mol. Biol. 346, 215–222 (2005).
Janssen, A., Beerling, E., Medema, R. & van Rheenen, J. Intravital FRET imaging of tumor cell viability and mitosis during chemotherapy. PLoS ONE 8, e64029 (2013).
Matsuda, T., Horikawa, K., Saito, K. & Nagai, T. Highlighted Ca2+ imaging with a genetically encoded 'caged' indicator. Sci. Rep. 3, 1398 (2013).
Demarco, I. A., Periasamy, A., Booker, C. F. & Day, R. N. Monitoring dynamic protein interactions with photoquenching FRET. Nature Methods 3, 519–524 (2006).
Subach, F. V. et al. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem. Biol. 17, 745–755 (2010).
Peter, M. et al. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 88, 1224–1237 (2005).
Talbot, C. B. et al. High speed unsupervised fluorescence lifetime imaging confocal multiwell plate reader for high content analysis. J. Biophoton. 1, 514–521 (2008). This study provides the first example of a FLIM biosensor incorporated into a high-throughput image-based screening platform.
Grecco, H. E. et al. In situ analysis of tyrosine phosphorylation networks by FLIM on cell arrays. Nature Methods 7, 467–472 (2010). This paper describes the elegant use of high-speed FLIM to measure FRET in a high-throughput setting, which provides insight into the concerted activity and network redundancy in epidermal growth factor receptor (EGFR) signalling. It is applicable to RTK-targeted drug resistance and feedback.
Kumar, S. et al. FLIM FRET technology for drug discovery: automated multiwell-plate high-content analysis, multiplexed readouts and application in situ. Chemphyschem 12, 609–626 (2011).
Alibhai, D. et al. Automated fluorescence lifetime imaging plate reader and its application to Forster resonant energy transfer readout of Gag protein aggregation. J. Biophoton. 6, 398–408 (2013).
Grecco, H. E., Roda-Navarro, P., Fengler, S. & Bastiaens, P. I. High-throughput quantification of posttranslational modifications in situ by CA-FLIM. Methods Enzymol. 500, 37–58 (2011).
McGhee, E. J. et al. FLIM-FRET imaging in vivo reveals 3D-environment spatially regulates RhoGTPase activity during cancer cell invasion. Small GTPases 2, 239–244 (2011).
Worth, D. C. & Parsons, M. Advances in imaging cell-matrix adhesions. J. Cell Sci. 123, 3629–3638 (2010).
Jares-Erijman, E. A. & Jovin, T. M. FRET imaging. Nature Biotech. 21, 1387–1395 (2003).
Berney, C. & Danuser, G. FRET or no FRET: a quantitative comparison. Biophys. J. 84, 3992–4010 (2003).
Roh-Johnson, M. et al. Macrophage contact induces RhoA GTPase signaling to trigger tumor cell intravasation. Oncogene http://dx.doi.org/10.1038/onc.2013.377 (2013).
Ottobrini, L., Martelli, C., Trabattoni, D. L., Clerici, M. & Lucignani, G. In vivo imaging of immune cell trafficking in cancer. Eur. J. Nucl. Med. Mol. Imag. 38, 949–968 (2011).
Chtanova, T. et al. Real-time interactive two-photon photoconversion of recirculating lymphocytes for discontinuous cell tracking in live adult mice. J. Biophoton http://dx.doi.org/10.1002/jbio.201200175 (2012).
Makrogianneli, K. et al. Integrating receptor signal inputs that influence small Rho GTPase activation dynamics at the immunological synapse. Mol. Cell. Biol. 29, 2997–3006 (2009).
Lohela, M. & Werb, Z. Intravital imaging of stromal cell dynamics in tumors. Curr. Opin. Genet. Dev. 20, 72–78 (2010).
Egeblad, M., Nakasone, E. S. & Werb, Z. Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell 18, 884–901 (2010).
Grant, D. M. et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys. J. 95, L69–L71 (2008).
Rao, J., Bhattacharya, D., Banerjee, B., Sarin, A. & Shivashankar, G. V. Trichostatin-A induces differential changes in histone protein dynamics and expression in HeLa cells. Biochem. Biophys. Res. Commun. 363, 263–268 (2007).
Li, W., Wang, Y., Shao, H., He, Y. & Ma, H. Probing rotation dynamics of biomolecules using polarization based fluorescence microscopy. Microsc. Res. Tech. 70, 390–395 (2007).
Cao, Z., Huang, C. C. & Tan, W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal. Chem. 78, 1478–1484 (2006).
Sharma, P. et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589 (2004).
Matthews, D. R. et al. A multi-functional imaging approach to high-content protein interaction screening. PLoS ONE 7, e33231 (2012).
Eichorst, J. P., Clegg, R. M. & Wang, Y. Red-shifted fluorescent proteins monitor enzymatic activity in live HT-1080 cells with fluorescence lifetime imaging microscopy (FLIM). J. Microsc. 248, 77–89 (2012).
Eichorst, J. P., Huang, H., Clegg, R. M. & Wang, Y. Phase differential enhancement of FLIM to distinguish FRET components of a biosensor for monitoring molecular activity of membrane type 1 matrix metalloproteinase in live cells. J. Fluoresc 21, 1763–1777 (2011).
Tyas, L., Brophy, V. A., Pope, A., Rivett, A. J. & Tavare, J. M. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 1, 266–270 (2000).
Shcherbakova, D. M., Hink, M. A., Joosen, L., Gadella, T. W. & Verkhusha, V. V. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J. Am. Chem. Soc. 134, 7913–7923 (2012).
Keese, M., Yagublu, V., Schwenke, K., Post, S. & Bastiaens, P. Fluorescence lifetime imaging microscopy of chemotherapy-induced apoptosis resistance in a syngenic mouse tumor model. Int. J. Cancer 126, 104–113 (2010).
Tomura, M. et al. Time-lapse observation of cellular function with fluorescent probe reveals novel CTL-target cell interactions. Int. Immunol. 21, 1145–1150 (2009).
Yamaguchi, Y. et al. Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J. Cell Biol. 195, 1047–1060 (2011).
Ting, A. Y., Kain, K. H., Klemke, R. L. & Tsien, R. Y. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl Acad. Sci. USA 98, 15003–15008 (2001).
Komatsu, N. et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656 (2011).
Kunkel, M. T., Ni, Q., Tsien, R. Y., Zhang, J. & Newton, A. C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 280, 5581–5587 (2005).
Sasaki, K., Sato, M. & Umezawa, Y. Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J. Biol. Chem. 278, 30945–30951 (2003).
Yoshizaki, H., Mochizuki, N., Gotoh, Y. & Matsuda, M. Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell 18, 119–128 (2007).
Mochizuki, N. et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411, 1065–1068 (2001).
Itoh, R. E. et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22, 6582–6591 (2002).
Kardash, E. et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nature Cell Biol. 12 (Suppl. 1–11), 47–53 (2010).
Timpson, P. et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 71, 747–757 (2011). This study demonstrates that subcellular FLIM–FRET imaging and targeting can reveal subtle but vital signalling events that drive tumour invasion in vivo.
Goto, A. et al. GDNF and endothelin 3 regulate migration of enteric neural crest-derived cells via protein kinase A and Rac1. J. Neurosci. 33, 4901–4912 (2013).
Johnsson, A. E. et al. The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Reports 6, 1153–1164 (2014). A RAC–FRET biosensor mouse was generated, allowing the spatiotemporal activity of RAC GTPase to be assessed in primary neutrophils and multiple organ types, such as the pancreas, liver, intestine and mammary tissue, in real-time. By crossing this inducible mouse with distinct tumour mouse models, we could expand our knowledge of how RAC GTPase behaves in a native host mammalian tissue upon drug treatment.
Fehr, M., Lalonde, S., Lager, I., Wolff, M. W. & Frommer, W. B. In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J. Biol. Chem. 278, 19127–19133 (2003).
Okumoto, S. et al. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl Acad. Sci. USA 102, 8740–8745 (2005).
Imamura, H. et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl Acad. Sci. USA 106, 15651–15656 (2009).
Ritsma, L., Vrisekoop, N. & van Rheenen, J. In vivo imaging and histochemistry are combined in the cryosection labelling and intravital microscopy technique. Nature Commun. 4, 2366 (2013). In this study, in a similar manner to electron microscopy, intravital images were correlated with cryosection labelling to provide post-imaging detail of the sample. Along with instant readouts from intravital imaging, this provides added contextual value and the long-term capacity to re-evaluate the sample after live imaging.
Potzkei, J. et al. Real-time determination of intracellular oxygen in bacteria using a genetically encoded FRET-based biosensor. BMC Biol. 10, 28 (2012). This paper describes the development of FluBO, an intramolecular FRET-based biosensor for detecting intracellular oxygen. FluBO uses an oxygen-insensitive donor fluorescent protein that is intramolecularly linked to an oxygen-sensitive acceptor fluorescent protein, and thus FRET only occurs in the presence of oxygen. This biosensor could eventually be applied to measure cellular hypoxia for in vivo cancer research.
Urra, J. et al. A genetically encoded ratiometric sensor to measure extracellular pH in microdomains bounded by basolateral membranes of epithelial cells. Pflugers Arch. 457, 233–242 (2008).
Awaji, T., Hirasawa, A., Shirakawa, H., Tsujimoto, G. & Miyazaki, S. Novel green fluorescent protein-based ratiometric indicators for monitoring pH in defined intracellular microdomains. Biochem. Biophys. Res. Commun. 289, 457–462 (2001).
Drepper, T. et al. Reporter proteins for in vivo fluorescence without oxygen. Nature Biotech. 25, 443–445 (2007).
Drepper, T. et al. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76, 5990–5994 (2010).
Piljic, A. & Schultz, C. Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 3, 156–160 (2008).
Peyker, A., Rocks, O. & Bastiaens, P. I. Imaging activation of two Ras isoforms simultaneously in a single cell. Chembiochem 6, 78–85 (2005).
Kamioka, Y. et al. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct. Funct. 37, 65–73 (2012). This paper describes the generation of FRET biosensor mice.
Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M. & Miyawaki, A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl Acad. Sci. USA 101, 10554–10559 (2004).
Atkin, S. D. et al. Transgenic mice expressing a cameleon fluorescent Ca2+ indicator in astrocytes and Schwann cells allow study of glial cell Ca2+ signals in situ and in vivo. J. Neurosci. Methods 181, 212–226 (2009).
Hara, M. et al. Imaging endoplasmic reticulum calcium with a fluorescent biosensor in transgenic mice. Am. J. Physiol. Cell Physiol. 287, C932–C938 (2004).
Isotani, E. et al. Real-time evaluation of myosin light chain kinase activation in smooth muscle tissues from a transgenic calmodulin-biosensor mouse. Proc. Natl Acad. Sci. USA 101, 6279–6284 (2004).
Bartoli, M. et al. A mouse model for monitoring calpain activity under physiological and pathological conditions. J. Biol. Chem. 281, 39672–39680 (2006).
Nikolaev, V. O., Bunemann, M., Schmitteckert, E., Lohse, M. J. & Engelhardt, S. Cyclic AMP imaging in adult cardiac myocytes reveals far-reaching β1-adrenergic but locally confined β2-adrenergic receptor-mediated signaling. Circ. Res. 99, 1084–1091 (2006).
Calebiro, D. et al. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 7, e1000172 (2009).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Yamamoto, N. et al. Cellular dynamics visualized in live cells in vitro and in vivo by differential dual-color nuclear-cytoplasmic fluorescent-protein expression. Cancer Res. 64, 4251–4256 (2004).
Yamamoto, N. et al. Determination of clonality of metastasis by cell-specific color-coded fluorescent-protein imaging. Cancer Res. 63, 7785–7790 (2003).
Hoffman, R. M. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nature Rev. Cancer 5, 796–806 (2005).
Day, R. N. & Davidson, M. W. Fluorescent proteins for FRET microscopy: monitoring protein interactions in living cells. Bioessays 34, 341–350 (2012).
Newman, R. H., Fosbrink, M. D. & Zhang, J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 111, 3614–3666 (2011).
Sahai, E. & Marshall, C. J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nature Cell Biol. 5, 711–719 (2003).
Friedl, P. & Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Rev. Cancer 3, 362–374 (2003).
Friedl, P., Sahai, E., Weiss, S. & Yamada, K. M. New dimensions in cell migration. Nature Rev. Mol. Cell. Biol. 13, 743–747 (2012). This paper gives a comprehensive insight into the appropriate use of 3D matrices to mimic in vivo homestasis or disease conditions.
Spence, H. J., Timpson, P., Tang, H. R., Insall, R. H. & Machesky, L. M. Scar/WAVE3 contributes to motility and plasticity of lamellipodial dynamics but not invasion in three dimensions. Biochem. J. 448, 35–42 (2012).
Wittig, R. et al. Biosensor-expressing spheroid cultures for imaging of drug-induced effects in three dimensions. J. Biomol. Screen 18, 736–743 (2013).
le Roux, L. et al. Optimizing imaging of three-dimensional multicellular tumor spheroids with fluorescent reporter proteins using confocal microscopy. Mol. Imag. 7, 214–221 (2008).
Uchugonova, A. et al. Multiphoton tomography visualizes collagen fibers in the tumor microenvironment that maintain cancer-cell anchorage and shape. J. Cell Biochem. 114, 99–102 (2013).
Pickl, M. & Ries, C. H. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 28, 461–468 (2009).
Calvo, F. et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nature Cell Biol. 15, 637–646 (2013).
Nurmenniemi, S. et al. A novel organotypic model mimics the tumor microenvironment. Am. J. Pathol. 175, 1281–1291 (2009).
Gaggioli, C. et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nature Cell Biol. 9, 1392–1400 (2007). This paper presents a strong argument for the use of organotypic co-culture models to investigate the complex systems involved in cancer metastasis.
Rothberg, J. M., Sameni, M., Moin, K. & Sloane, B. F. Live-cell imaging of tumor proteolysis: impact of cellular and non-cellular microenvironment. Biochim. Biophys. Acta 1824, 123–132 (2012).
Wang, R. et al. Three-dimensional co-culture models to study prostate cancer growth, progression, and metastasis to bone. Semin. Cancer Biol. 15, 353–364 (2005).
Talukdar, S. & Kundu, S. C. A. non-mulberry silk fibroin protein based 3D in vitro tumor model for evaluation of anticancer drug activity. Adv. Funct. Mater. 22, 4778–4788 (2012).
Serebriiskii, I., Castello-Cros, R., Lamb, A., Golemis, E. A. & Cukierman, E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 27, 573–585 (2008).
Loessner, D. et al. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31, 8494–8506 (2010).
Schrader, J. et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 53, 1192–1205 (2011).
Sethi, T. et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nature Med. 5, 662–668 (1999).
Longati, P. et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 13, 95 (2013).
Straussman, R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012). This study demonstrated that stromal cells conferred innate chemoresistance to cancer cells through treatment with an array of 35 anticancer drugs in 45 cancer cells, cultured alone or in co-culture with stromal cells.
Levental, K. R. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Mih, J. D., Marinkovic, A., Liu, F., Sharif, A. S. & Tschumperlin, D. J. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J. Cell Sci. 125, 5974–5983 (2012).
Zustiak, S., Nossal, R. & Sackett, D. L. Multiwell stiffness assay for the study of cell responsiveness to cytotoxic drugs. Biotechnol. Bioeng. 111, 396–403 (2013).
Wolf, K. et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature Cell Biol. 9, 893–904 (2007).
Tozluoglu, M. et al. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nature Cell Biol. 15, 751–762 (2013).
Boghaert, E. et al. Host epithelial geometry regulates breast cancer cell invasiveness. Proc. Natl Acad. Sci. USA 109, 19632–19637 (2012).
Radisky, D. C. & Nelson, C. M. Regulation of mechanical stress by mammary epithelial tissue structure controls breast cancer cell invasion. Oncotarget 4, 498–499 (2013).
Jacobetz, M. A. et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 62, 112–120 (2013).
Olive, K. P. et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324, 1457–1461 (2009). In this study, SHG imaging reveals that targeting the tumour stromal compartment (ECM) before drug treatment can enhance drug delivery and improve disease outcome. This study led to a shift in the field of dual-targeted therapy in pancreatic cancer.
Provenzano, P. P. et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21, 418–429 (2012).
Yu, M. & Tannock, I. F. Targeting tumor architecture to favor drug penetration: a new weapon to combat chemoresistance in pancreatic cancer? Cancer Cell 21, 327–329 (2012). References 169 and 172 reveal that alterations in ECM integrity can influence drug targeting via reduced ECM content and improved vascularity or drug delivery.
Raub, C. B., Putnam, A. J., Tromberg, B. J. & George, S. C. Predicting bulk mechanical properties of cellularized collagen gels using multiphoton microscopy. Acta Biomater. 6, 4657–4665 (2010).
Samuel, M. S. et al. Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell 19, 776–791 (2011).
Cicchi, R. et al. Scoring of collagen organization in healthy and diseased human dermis by multiphoton microscopy. J. Biophoton. 3, 34–43 (2010).
Bakker, G. J., Andresen, V., Hoffman, R. M. & Friedl, P. Fluorescence lifetime microscopy of tumor cell invasion, drug delivery, and cytotoxicity. Methods Enzymol. 504, 109–125 (2012). This paper highlights the feasibility of high-speed FLIM–FRET in 3D organotypic or complex settings, allowing cancer cell invasion, apoptosis and drug uptake to be quantified with subcellular resolution.
Bremer, C., Tung, C. H. & Weissleder, R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nature Med. 7, 743–748 (2001).
Contag, P. R. Whole-animal cellular and molecular imaging to accelerate drug development. Drug Discov. Today 7, 555–562 (2002).
Graves, E. E., Weissleder, R. & Ntziachristos, V. Fluorescence molecular imaging of small animal tumor models. Curr. Mol. Med. 4, 419–430 (2004).
Hoffman, R. M. & Yang, M. Whole-body imaging with fluorescent proteins. Nature Protoc. 1, 1429–1438 (2006).
Hayashi, K. et al. Real-time imaging of tumor-cell shedding and trafficking in lymphatic channels. Cancer Res. 67, 8223–8228 (2007).
Yamauchi, K. et al. Development of real-time subcellular dynamic multicolor imaging of cancer-cell trafficking in live mice with a variable-magnification whole-mouse imaging system. Cancer Res. 66, 4208–4214 (2006).
Yang, M. et al. Direct external imaging of nascent cancer, tumor progression, angiogenesis, and metastasis on internal organs in the fluorescent orthotopic model. Proc. Natl Acad. Sci. USA 99, 3824–3829 (2002).
Giampieri, S. et al. Localized and reversible TGFβ signalling switches breast cancer cells from cohesive to single cell motility. Nature Cell Biol. 11, 1287–1296 (2009).
Li, A. et al. Rac1 drives melanoblast organization during mouse development by orchestrating pseudopod- driven motility and cell-cycle progression. Dev. Cell 21, 722–734 (2011).
Ladhani, O., Sanchez-Martinez, C., Orgaz, J. L., Jimenez, B. & Volpert, O. V. Pigment epithelium-derived factor blocks tumor extravasation by suppressing amoeboid morphology and mesenchymal proteolysis. Neoplasia 13, 633–642 (2011).
Patsialou, A. et al. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. IntraVital 2, e25294 (2013).
Ahmed, F. et al. GFP expression in the mammary gland for imaging of mammary tumor cells in transgenic mice. Cancer Res. 62, 7166–7169 (2002).
Zomer, A. et al. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31, 602–606 (2013). This paper describes the elegant use of simultaneous cell tagging to track cell fate and clonal progeny, and this approach was subsequently used in combination with imaging windows to monitor cell fate in tumour tissue.
Barretto, R. P. et al. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nature Med. 17, 223–228 (2011).
Tang, J. C. et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 154, 928–939 (2013).
Momiyama, M. et al. Imaging the efficacy of UVC irradiation on superficial brain tumors and metastasis in live mice at the subcellular level. J. Cell Biochem. 114, 428–434 (2013).
Morton, J. P. et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl Acad. Sci. USA 107, 246–251 (2010). This paper describes how whole-body imaging and single cell tagging revealed a role for p53 in outgrowth of senescence, which resulted in tumour progression.
Morton, J. P. et al. Dasatinib inhibits the development of metastases in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 139, 292–303 (2010).
Ahmad, I. et al. β-Catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene 30, 178–189 (2011).
Cole, A. M. et al. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO Mol. Med. 2, 472–486 (2010).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Roth, S. et al. Paneth cells in intestinal homeostasis and tissue injury. PLoS ONE 7, e38965 (2012).
Myant, K. B. et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 12, 761–773 (2013).
Chan, K. T. et al. Intravital imaging of a spheroid-based orthotopic model of melanoma in the mouse ear skin. IntraVital 2, e25805 (2013).
Lindsay, C. R. et al. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nature Commun. 2, 555 (2011).
Doyle, B. et al. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J. Pathol. 222, 129–137 (2010).
Gritsenko, P. G., Ilina, O. & Friedl, P. Interstitial guidance of cancer invasion. J. Pathol. 226, 185–199 (2012).
Dirat, B. et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 71, 2455–2465 (2011).
Nieman, K. M. et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature Med. 17, 1498–1503 (2011).
Wolf, K. et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 (2013).
Le Devedec, S. E. et al. Two-photon intravital multicolour imaging to study metastatic behaviour of cancer cells in vivo. Methods Mol. Biol. 769, 331–349 (2011).
Coffey, S. E., Giedt, R. J. & Weissleder, R. Automated analysis of clonal cancer cells by intravital imaging. IntraVital 2, e26138 (2013).
Wyckoff, J. et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 64, 7022–7029 (2004).
Wyckoff, J. B. et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67, 2649–2656 (2007).
Lin, E. Y. et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 66, 11238–11246 (2006).
Zhou, Z. N. et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene http://dx.doi.org/10.1038/onc.2013.363 (2013).
Xu, Z. et al. Role of pancreatic stellate cells in pancreatic cancer metastasis. Am. J. Pathol. 177, 2585–2596 (2010).
Brown, E. et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nature Med. 9, 796–800 (2003).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nature Med. 16, 116–122 (2010). This study demonstrates the use of CIW technology to track metastatic site colonization of cancer cells.
Gligorijevic, B. & Condeelis, J. Stretching the timescale of intravital imaging in tumors. Cell Adh Migr. 3, 313–315 (2009).
Gligorijevic, B., Kedrin, D., Segall, J. E., Condeelis, J. & van Rheenen, J. Dendra2 photoswitching through the mammary imaging window. J. Vis. Exp. 28, 1278 (2009).
Ritsma, L. et al. Surgical implantation of an abdominal imaging window for intravital microscopy. Nature Protoc. 8, 583–594 (2013). This paper describes the application of AIWs to repeatedly and non-invasively monitor organs from deep within the abdominal cavity. Liver, pancreas, intestine and kidney were imaged at high resolution in situ.
Giedt, R. J., Koch, P. D. & Weissleder, R. Single cell analysis of drug distribution by intravital imaging. PLoS ONE 8, e60988 (2013).
Agasti, S. S., Laughney, A. M., Kohler, R. H. & Weissleder, R. A photoactivatable drug-caged fluorophore conjugate allows direct quantification of intracellular drug transport. Chem. Commun. 49, 11050–11052 (2013).
Thurber, G. M. et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nature Commun. 4, 1504 (2013). In this study, the authors use intravital imaging to monitor a PARP1 inhibitor reaching its target compartment within the cell in vivo.
US Department of Health and Human Services. Food and Drug Administration (FDA) Centre for Drug Evaluation and Research (CDER). Guidance for Industry: Codevelopment of two or more new investigational drugs for use in combination. June 2013. Clinical Medicine.
Hughes, B. Novel agents combined get own guidance. Nature Biotech. 29, 174 (2011).
Natale, D., Soriano, S. F., Coelho, F. M., Hons, M. & Stein, J. V. Comprehensive assessment of quantum dots for multispectral twophoton imaging of dynamic leukocyte migration in lymph nodes. IntraVital 2, e25745 (2013).
Suetsugu, A. et al. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev. 65, 383–390 (2013).
Chaudhry, S. I. et al. Autocrine IL-1β-TRAF6 signalling promotes squamous cell carcinoma invasion through paracrine TNFα signalling to carcinoma-associated fibroblasts. Oncogene 32, 747–758 (2013).
Zhang, J. & Liu, J. Tumor stroma as targets for cancer therapy. Pharmacol. Ther. 137, 200–215 (2013).
Bousso, P. & Moreau, H. D. Functional immunoimaging: the revolution continues. Nature Rev. Immunol. 12, 858–864 (2012).
Carmeliet, P. & Jain, R. K. Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 (2011).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Clarke, J. M. & Hurwitz, H. I. Targeted inhibition of VEGF receptor 2: an update on ramucirumab. Expert Opin. Biol. Ther. 13, 1187–1196 (2013).
Di Marco, M., Macchini, M., Vecchiarelli, S., Sina, S. & Biasco, G. Hedgehog signaling: from the cuirass to the heart of pancreatic cancer. Pancreatology 12, 388–393 (2012).
Hoffman, R. M. & Yang, M. Color-coded fluorescence imaging of tumor-host interactions. Nature Protoc. 1, 928–935 (2006).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Amoh, Y., Li, L., Katsuoka, K., Bouvet, M. & Hoffman, R. M. GFP-expressing vascularization of Gelfoam as a rapid in vivo assay of angiogenesis stimulators and inhibitors. Biotechniques 42, 294–298 (2007).
Tanaka, K. et al. In vivo real-time imaging of chemotherapy response on the liver metastatic tumor microenvironment using multiphoton microscopy. Oncol. Rep. 28, 1822–1830 (2012).
Manning, C. S. et al. Intravital imaging reveals conversion between distinct tumor vascular morphologies and localized vascular response to Sunitinib. IntraVital 2, e24790 (2013).
Olivier, N. et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329, 967–971 (2010).
Weigelin, B., Bakker, G. & Friedl, P. Intravital third harmonic generation microscopy of collective melanoma cell invasion: Principles of interface guidance and microvesicle dynamics. IntraVital 1, 32–43 (2012).
Peinado, H. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 18, 883–891 (2012).
Tadrous, P. J. et al. Fluorescence lifetime imaging of unstained tissues: early results in human breast cancer. J. Pathol. 199, 309–317 (2003).
Provenzano, P. P., Eliceiri, K. W. & Keely, P. J. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin. Exp. Metastasis 26, 357–370 (2009).
McGinty, J. et al. Wide-field fluorescence lifetime imaging of cancer. Biomed. Opt. Express 1, 627–640 (2010).
Estrella, V. et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 73, 1524–1535 (2013).
Skala, M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl Acad. Sci. USA 104, 19494–19499 (2007).
Lecoq, J. et al. Simultaneous two-photon imaging of oxygen and blood flow in deep cerebral vessels. Nature Med. 17, 893–898 (2011).
Belousov, V. V. et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nature Methods 3, 281–286 (2006).
Parpaleix, A., Houssen, Y. G. & Charpak, S. Imaging local neuronal activity by monitoring PO2 transients in capillaries. Nature Med. 19, 241–246 (2013).
Timpson, P. et al. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. J. Vis. Exp. 56, e3089 (2011).
Thoma, C. R. et al. A high-throughput-compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. J. Biomol. Screen 18, 1330–1337 (2013).
Tung, Y. C. et al. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 136, 473–478 (2011).
Drewitz, M. et al. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 6, 1488–1496 (2011).
Burgstaller, G., Oehrle, B., Koch, I., Lindner, M. & Eickelberg, O. Multiplex profiling of cellular invasion in 3D cell culture models. PLoS ONE 8, e63121 (2013).
Truong, H. H. et al. Automated microinjection of cell-polymer suspensions in 3D ECM scaffolds for high-throughput quantitative cancer invasion screens. Biomaterials 33, 181–188 (2012).
Zervantonakis, I. K. et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl Acad. Sci. USA 109, 13515–13520 (2012).
Echeverria, V. et al. An automated high-content assay for tumor cell migration through 3-dimensional matrices. J. Biomol. Screen 15, 1144–1151 (2010).
Kim, J. et al. A programmable microfluidic cell array for combinatorial drug screening. Lab. Chip 12, 1813–1822 (2012).
Alencar, H., Mahmood, U., Kawano, Y., Hirata, T. & Weissleder, R. Novel multiwavelength microscopic scanner for mouse imaging. Neoplasia 7, 977–983 (2005).
Al-Gubory, K. H. & Houdebine, L. M. In vivo imaging of green fluorescent protein-expressing cells in transgenic animals using fibred confocal fluorescence microscopy. Eur. J. Cell Biol. 85, 837–845 (2006).
Kennedy, G. T. et al. A fluorescence lifetime imaging scanning confocal endomicroscope. J. Biophoton. 3, 103–107 (2010).
Kiesslich, R. et al. Identification of epithelial gaps in human small and large intestine by confocal endomicroscopy. Gastroenterology 133, 1769–1778 (2007).
Stallmach, A., Schmidt, C., Watson, A. & Kiesslich, R. An unmet medical need: advances in endoscopic imaging of colorectal neoplasia. J. Biophoton. 4, 482–489 (2011).
Dancik, Y., Favre, A., Loy, C. J., Zvyagin, A. V. & Roberts, M. S. Use of multiphoton tomography and fluorescence lifetime imaging to investigate skin pigmentation in vivo. J. Biomed. Opt. 18, 26022 (2013).
Leite-Silva, V. R. et al. The effect of formulation on the penetration of coated and uncoated zinc oxide nanoparticles into the viable epidermis of human skin in vivo. Eur. J. Pharm. Biopharm. 84, 297–308 (2013).
Sanchez, W. Y., Obispo, C., Ryan, E., Grice, J. E. & Roberts, M. S. Changes in the redox state and endogenous fluorescence of in vivo human skin due to intrinsic and photo-aging, measured by multiphoton tomography with fluorescence lifetime imaging. J. Biomed. Opt. 18, 061217 (2013).
Carragher, N. O. & Frame, M. C. Modelling distinct modes of tumour invasion and metastasis. Drug Discov. Today Dis. Models 8, 103–112 (2011).
Sameni, M. et al. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin. Exp. Metastasis 26, 299–309 (2009).
Carragher, N. O. Profiling distinct mechanisms of tumour invasion for drug discovery: imaging adhesion, signalling and matrix turnover. Clin. Exp. Metastasis 26, 381–397 (2009).
Boimel, P. J. et al. Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 14, R23 (2012).
Xue, C. et al. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 66, 192–197 (2006).
Ai, H. W., Henderson, J. N., Remington, S. J. & Campbell, R. E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400, 531–540 (2006).
Markwardt, M. L. et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS ONE 6, e17896 (2011).
Goedhart, J. et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nature Methods 7, 137–139 (2010).
Goedhart, J. et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nature Commun. 3, 751 (2012).
Goedhart, J., Vermeer, J. E., Adjobo-Hermans, M. J., van Weeren, L. & Gadella, T. W. Jr. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE 2, e1011 (2007).
Aoki, K., Kamioka, Y. & Matsuda, M. Fluorescence resonance energy transfer imaging of cell signaling from in vitro to in vivo: basis of biosensor construction, live imaging, and image processing. Dev. Growth Differ. 55, 515–522 (2013).
Truong, K. et al. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nature Struct. Biol. 8, 1069–1073 (2001).
Miyawaki, A. et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997).
Miyawaki, A., Griesbeck, O., Heim, R. & Tsien, R. Y. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc. Natl Acad. Sci. USA 96, 2135–2140 (1999).
Griesbeck, O., Baird, G. S., Campbell, R. E., Zacharias, D. A. & Tsien, R. Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276, 29188–29194 (2001).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotech. 20, 87–90 (2002).
Evanko, D. S. & Haydon, P. G. Elimination of environmental sensitivity in a cameleon FRET-based calcium sensor via replacement of the acceptor with Venus. Cell Calcium 37, 341–348 (2005).
Vinkenborg, J. L., Evers, T. H., Reulen, S. W., Meijer, E. W. & Merkx, M. Enhanced sensitivity of FRET-based protease sensors by redesign of the GFP dimerization interface. Chembiochem 8, 1119–1121 (2007).
Ohashi, T., Galiacy, S. D., Briscoe, G. & Erickson, H. P. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 16, 1429–1438 (2007).
Nguyen, A. W. & Daugherty, P. S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nature Biotech. 23, 355–360 (2005).
Lam, A. J. et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nature Methods 9, 1005–1012 (2012).
Fritz, R. D. et al. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci Signal 6, rs12 (2013).
Golynskiy, M. V., Rurup, W. F. & Merkx, M. Antibody detection by using a FRET-based protein conformational switch. Chembiochem 11, 2264–2267 (2010).
Rehm, M. et al. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J. Biol. Chem. 277, 24506–24514 (2002).
Takemoto, K., Nagai, T., Miyawaki, A. & Miura, M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160, 235–243 (2003).
Onuki, R. et al. Confirmation by FRET in individual living cells of the absence of significant amyloid β-mediated caspase 8 activation. Proc. Natl Acad. Sci. USA 99, 14716–14721 (2002).
Li, M., Chen, X., Ye, Q. Z., Vogt, A. & Yin, X. M. A high-throughput FRET-based assay for determination of Atg4 activity. Autophagy 8, 401–412 (2012).
Macurek, L. et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 (2008).
Harvey, C. D. et al. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl Acad. Sci. USA 105, 19264–19269 (2008).
Mizutani, T. et al. A novel FRET-based biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clin. Cancer Res. 16, 3964–3975 (2010).
Randriamampita, C. et al. A novel ZAP-70 dependent FRET based biosensor reveals kinase activity at both the immunological synapse and the antisynapse. PLoS ONE 3, e1521 (2008).
Acknowledgements
The authors thank H. Bennett, D. Herrmann, A. Magenau, A. Burgess, M. Pajic, B. Browne and C. Vennin. This work was supported by the Australian Research Council (ARC), Cancer Institute New South Wales (CINSW) and National Health and Medical Research Council (NHMRC) funding. N.O.C. is supported by a Research Councils United Kingdom (RCUK) fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Phenotypic screening
-
Assay systems that enable quantifiable measurements of cell phenotype or function that can be used to guide compound selection or iterative chemical design, often in the absence of any prior knowledge of an intended drug target.
- First-in-class small molecule medicines
-
Newly approved medicines that have novel mechanisms of action, distinct from anything else on the market.
- Target-directed drug discovery
-
A contemporary strategy for the identification and optimization of lead molecules and candidate drugs based on achieving high levels of potency and specificity against a nominated target that is implicated in disease progression.
- Anisotropy
-
The anisotropy of a molecule is assessed through the simultaneous measurement of orthogonally polarized fluorescence relative to the polarization of the excitation light. Factors that determine the degree of anisotropy are protein mobility and molecular orientation. As a consequence, anisotropy can be used as a powerful and sensitive readout for binding and screening assays of protein behaviour and interactions.
- Extracellular matrix
-
(ECM). A reinforced composite of structural proteins that is primarily composed of collagen and tissue-specific inclusions (for example, fibronectin and laminin), as well as other metabolites secreted by cells. The ECM provides structural support and biochemical signals for multicellular tissue and organ systems.
- Intravital imaging windows
-
Windows that are surgically implanted in a mouse to allow repeated, non-invasive imaging over a long time course.
- Organotypic 3D collagen I matrix
-
Fibroblast-driven contraction of acid-extracted collagen I is used to produce matrices with high in vivo fidelity for analysis of cell behaviour in a live in vitro setting.
- Multiphoton intravital microscopy
-
This method reduces interference from the background by using more than one photon as a multiple of the excitation wavelength of the sample, effectively restricting interactions to the focal plane and allowing deep imaging within live tissue.
Rights and permissions
About this article
Cite this article
Conway, J., Carragher, N. & Timpson, P. Developments in preclinical cancer imaging: innovating the discovery of therapeutics. Nat Rev Cancer 14, 314–328 (2014). https://doi.org/10.1038/nrc3724
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3724
This article is cited by
-
Multiphoton intravital microscopy of rodents
Nature Reviews Methods Primers (2022)
-
Spatial heterogeneity of oxygenation and haemodynamics in breast cancer resolved in vivo by conical multispectral optoacoustic mesoscopy
Light: Science & Applications (2020)
-
Application of machine learning method in optical molecular imaging: a review
Science China Information Sciences (2020)
-
Real-time in vivo imaging of subpopulations of circulating tumor cells using antibody conjugated quantum dots
Journal of Nanobiotechnology (2019)
-
The serum amyloid A3 promoter-driven luciferase reporter mice is a valuable tool to image early renal fibrosis development and shows the therapeutic effect of glucosyl-hesperidin treatment
Scientific Reports (2019)