Key Points
-
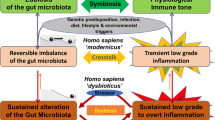
A narrow definition of dysbiosis is as a stable microbial community state that functionally contributes to the aetiology, diagnosis or treatment of a disease.
-
Dysbiosis is often driven by infection and inflammation, diet and xenobiotics, host genetics or the host's environment.
-
Innate and adaptive immunity control the colonization niche of the intestinal microbiota through mechanisms including the production of antimicrobial peptides and IgA antibodies.
-
A dysbiotic microbiota may actively influence its colonization niche by altering the functions of innate and adaptive intestinal immunity.
-
Dysbiosis has been associated with many immune-related human diseases, but in many cases it remains to be established whether dysbiosis is a cause or consequence of the disease.
-
Personalized nutrition and metabolite-based 'postbiotic' therapy may present ways in which to harness the increasing knowledge about dysbiosis in disease for the design of new therapies.
Abstract
Throughout the past century, we have seen the emergence of a large number of multifactorial diseases, including inflammatory, autoimmune, metabolic, neoplastic and neurodegenerative diseases, many of which have been recently associated with intestinal dysbiosis — that is, compositional and functional alterations of the gut microbiome. In linking the pathogenesis of common diseases to dysbiosis, the microbiome field is challenged to decipher the mechanisms involved in the de novo generation and the persistence of dysbiotic microbiome configurations, and to differentiate causal host–microbiome associations from secondary microbial changes that accompany disease course. In this Review, we categorize dysbiosis in conceptual terms and provide an overview of immunological associations; the causes and consequences of bacterial dysbiosis, and their involvement in the molecular aetiology of common diseases; and implications for the rational design of new therapeutic approaches. A molecular- level understanding of the origins of dysbiosis, its endogenous and environmental regulatory processes, and its downstream effects may enable us to develop microbiome-targeting therapies for a multitude of common immune-mediated diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bach, J. F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347, 911–920 (2002).
Egger, G. & Dixon, J. Beyond obesity and lifestyle: a review of 21st century chronic disease determinants. Biomed Res. Int. 2014, 731685 (2014).
Kelsen, J. R. & Wu, G. D. The gut microbiota, environment and diseases of modern society. Gut Microbes 3, 374–382 (2012).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). This study highlights extensive interindividual variability in the composition of the microbiome of healthy individuals.
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome Med. 8, 51 (2016).
Stappenbeck, T. S. & Virgin, H. W. Accounting for reciprocal host−microbiome interactions in experimental science. Nature 534, 191–199 (2016).
Thaiss, C. A. et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551 (2016).
Fonseca, D. M. et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell 163, 354–366 (2015). This paper describes a role for the microbiome in mediating post-infection immune system aberrations.
Chow, J. & Mazmanian, S. K. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276 (2010).
Stecher, B., Maier, L. & Hardt, W. D. 'Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11, 277–284 (2013).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007). This study describes microbially transmissible colitis in mice that lack T-bet in the innate immune system.
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Buffington, S. A. et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775 (2016).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). References 18–20 describe how microbiome reconstitution approaches can be used to treat dysbiosis-associated phenotypes.
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Erny, D. et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015).
Levy, M. et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 (2015). This study demonstrates that microbial hijacking of the innate immune system contributes to the establishment of dysbiosis.
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167, 1469–1480.e12 (2016).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013). References 25 and 26 report that the decrease in microbiome richness predisposes patients to dysmetabolism and low-grade inflammation.
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015).
Monaco, C. L. et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19, 311–322 (2016).
Kostic, A. D. et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273 (2015).
Mosca, A., Leclerc, M. & Hugot, J. P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front. Microbiol. 7, 455 (2016).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007).
Stecher, B. et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Ayres, J. S., Trinidad, N. J. & Vance, R. E. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18, 799–806 (2012).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013).
Deriu, E. et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013).
Stecher, B. et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl Acad. Sci. USA 109, 1269–1274 (2012).
Behnsen, J. et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273 (2014).
Lopez, C. A. et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Denou, E., Marcinko, K., Surette, M. G., Steinberg, G. R. & Schertzer, J. D. High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 310, E982–E993 (2016).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Suez, J. et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186 (2014).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
Levy, M., Thaiss, C. A. & Elinav, E. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 7, 120 (2015).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Bonder, M. J. et al. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016).
Turpin, W. et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016).
Wang, J. et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406 (2016). References 49–52 highlight the influence of host genetics on microbiota composition.
Benson, A. K. et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18933–18938 (2010).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
McCafferty, J. et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 7, 2116–2125 (2013).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Thaiss, C. A. et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014).
Voigt, R. M. et al. The circadian clock mutation promotes intestinal dysbiosis. Alcohol. Clin. Exp. Res. 40, 335–347 (2016).
Koren, O. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480 (2012).
Kigerl, K. A. et al. Gut dysbiosis impairs recovery after spinal cord injury. J. Exp. Med. 213, 2603–2620 (2016).
Thaiss, C. A., Levy, M., Suez, J. & Elinav, E. The interplay between the innate immune system and the microbiota. Curr. Opin. Immunol. 26, 41–48 (2014).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008).
Frantz, A. L. et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 5, 501–512 (2012).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010). The authors of this study report microbial transmissible metabolic syndrome in mice that lack TLR5.
Carvalho, F. A. et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12, 139–152 (2012).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013).
Petnicki-Ocwieja, T. et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl Acad. Sci. USA 106, 15813–15818 (2009).
Rehman, A. et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60, 1354–1362 (2011).
Li, E. et al. Inflammatory bowel diseases phenotype. C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS ONE 7, e26284 (2012).
Robertson, S. J. et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 4, 222–231 (2013).
Henao-Mejia, J., Elinav, E., Thaiss, C. A. & Flavell, R. A. Inflammasomes and metabolic disease. Annu. Rev. Physiol. 76, 57–78 (2014).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Hu, B. et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl Acad. Sci. USA 110, 9862–9867 (2013).
Wlodarska, M. et al. NLRP6 inflammasome orchestrates the colonic host–microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059 (2014).
Birchenough, G. M., Nystrom, E. E., Johansson, M. E. & Hansson, G. C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016).
Rausch, P. et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 306, 343–355 (2016).
Chudnovskiy, A. et al. Host–protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–456.e14 (2016).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13 (2016).
Sonnenberg, G. F. & Artis, D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity 37, 601–610 (2012).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Qiu, J. et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 39, 386–399 (2013).
Sonnenberg, G. F. et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012).
Powell, N. et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity 37, 674–684 (2012).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Cullender, T. C. et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581 (2013).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Shulzhenko, N. et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 17, 1585–1593 (2011).
Fagarasan, S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004).
Kawamoto, S. et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336, 485–489 (2012).
Nieuwenhuis, E. E. et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J. Clin. Invest. 119, 1241–1250 (2009).
Ismail, A. S. et al. γδ intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc. Natl Acad. Sci. USA 108, 8743–8748 (2011).
Thaiss, C. A., Zmora, N., Levy, M. & Elinav, E. The microbiome and innate immunity. Nature 535, 65–74 (2016).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016). This study demonstrates that variations in the immunostimulatory activity of LPS in different microbiomes are associated with the development of autoimmune diseases in children.
Maekawa, T. et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15, 768–778 (2014).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Zenewicz, L. A. et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190, 5306–5312 (2013).
Moon, C. et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521, 90–93 (2015). This paper shows that commensal bacteria modulate intestinal antibody levels via secretory IgA degradation.
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014).
Strauss, J. et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978 (2011).
Li, J., Butcher, J., Mack, D. & Stintzi, A. Functional impacts of the intestinal microbiome in the pathogenesis of inflammatory bowel disease. Inflamm. Bowel Dis. 21, 139–153 (2015).
Rigottier-Gois, L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 7, 1256–1261 (2013).
Wacklin, P. et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 19, 934–941 (2013).
Jabri, B. & Sollid, L. M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 9, 858–870 (2009).
Di Cagno, R. et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 11, 219 (2011).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015).
Achenbach, P., Bonifacio, E., Koczwara, K. & Ziegler, A. G. Natural history of type 1 diabetes. Diabetes 54 (Suppl. 2), S25–S31 (2005).
Risnes, K. R., Belanger, K., Murk, W. & Bracken, M. B. Antibiotic exposure by 6 months and asthma and allergy at 6 years: findings in a cohort of 1,401 US children. Am. J. Epidemiol. 173, 310–318 (2011).
Russell, S. L. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012).
Hill, D. A. et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18, 538–546 (2012).
Cahenzli, J., Koller, Y., Wyss, M., Geuking, M. B. & McCoy, K. D. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 14, 559–570 (2013).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Bisgaard, H. et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 357, 1487–1495 (2007).
Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl Med. 7, 307ra152 (2015).
Chen, J. et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484 (2016).
Berer, K. et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011).
Marchesi, J. R. et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 6, 546–551 (2007).
Olesen, S. W. & Alm, E. J. Dysbiosis is not an answer. Nat. Microbiol. 1, 16228 (2016).
Scheperjans, F. et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov. Disord. 30, 350–358 (2015).
Minter, M. R. et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer's disease. Sci. Rep. 6, 30028 (2016).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Angelberger, S. et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am. J. Gastroenterol. 108, 1620–1630 (2013).
Khan, K. J. et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am. J. Gastroenterol. 106, 661–673 (2011).
Wolvers, D. et al. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of infections by probiotics. J. Nutr. 140, 698S–712S (2010).
Maldonado-Gomez, M. X. et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20, 515–526 (2016).
Zmora, N., Zeevi, D., Korem, T., Segal, E. & Elinav, E. Taking it personally: personalized utilization of the human microbiome in health and disease. Cell Host Microbe 19, 12–20 (2016).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Wang, Z. et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Viaud, S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).
Vétizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Thaiss, C. A. & Elinav, E. Exploring new horizons in microbiome research. Cell Host Microbe 15, 662–667 (2014).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Chu, H. et al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Tsilingiri, K. et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut 61, 1007–1015 (2012).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Levy, M., Thaiss, C. A. & Elinav, E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. 30, 1589–1597 (2016).
Chang, P. V., Hao, L., Offermanns, S. & Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA 111, 2247–2252 (2014).
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Gomez de Aguero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Atarashi, K. et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015).
Sano, T. et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393 (2015).
Lecuyer, E. et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620 (2014).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Bunker, J. J. et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015).
Jiang, W. et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 5, 8096 (2015).
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Vrieze, A. et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7 (2012).
Reijnders, D. et al. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 24, 63–74 (2016).
Caesar, R., Tremaroli, V., Kovatcheva-Datchary, P., Cani, P. D. & Backhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22, 658–668 (2015).
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012).
Dapito, D. H. et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516 (2012).
Choi, G. B. et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016).
Finegold, S. M. et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 16, 444–453 (2010).
Sandler, R. H. et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J. Child Neurol. 15, 429–435 (2000).
Acknowledgements
The authors thank the members of the Elinav laboratory for fruitful discussions. They apologize to those authors whose work could not be cited owing to space limitations. A.A.K. is supported by a European Molecular Biology Organization postdoctoral fellowship. C.A.T. received a Boehringer Ingelheim Fonds Ph.D. Fellowship. E.E. is supported by Yael and Rami Ungar, Israel; the Leona M. and Harry B. Helmsley Charitable Trust; the Gurwin Family Fund for Scientific Research; the Crown Endowment Fund for Immunological Research; the estate of Jack Gitlitz; the estate of Lydia Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; the Adelis Foundation; John L. and Vera Schwartz, Pacific Palisades, California, USA; Alan Markovitz, Canada; Cynthia Adelson, Canada; Centre National de la Recherche Scientifique; the estate of Samuel and Alwyn J. Weber; Mr and Mrs Schwarz, Sherman Oaks, California, USA; grants funded by the European Research Council; the German–Israel Binational foundation; the Israel Science Foundation; the Minerva Foundation; the Rising Tide Foundation; and the Alon Foundation scholar award. E.E. is the incumbent of the Rina Gudinski Career Development Chair and a senior fellow of the Canadian Institute For Advanced Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Xenobiotics
-
Small chemical compounds that enter an organism unnaturally, such as drugs or pollutants.
- Koch's postulates
-
A list of criteria that a microorganism needs to fulfil to be considered the causative agent of a disease, including its presence in all cases of the disease, the ability to grow the microorganism in pure culture, transmissibility of the disease by inoculation of a healthy organism and the re-isolation of the microorganism from the infected host.
- Alpha diversity
-
Alpha diversity describes species richness within a site, in contrast to beta diversity, which refers to differences in species composition between sites.
- Antimicrobial peptides
-
Host-derived peptides that are part of the innate immune system and function in host defence against microorganisms.
- Metabolic syndrome
-
A syndrome of co-occurring conditions, including elevated levels of plasma glucose, high blood pressure, abdominal obesity, elevated serum triglyceride levels and low serum high-density lipoprotein levels, that collectively increase the risk of diabetes, stroke and heart disease.
- Inflammasomes
-
Multiprotein complexes composed of a NOD-like receptor protein, the adaptor protein ASC and caspase 1. Inflammasomes contribute to the secretion of interleukin-1β (IL-1β) and IL-18 by activating caspase 1.
- Innate lymphoid cells
-
(ILCs). Cells of the lymphoid lineage that do not express antigen-specific receptors, but orchestrate tissue homeostasis and immunity through cytokine secretion.
- Metabolome
-
The entirety of small-molecule metabolites at a particular site.
- Probiotics
-
Microorganisms that are administered to an organism to benefit the host.
- Prebiotics
-
Ingredients of food that promote the growth and metabolism of beneficial microorganisms in the host.
- Human microbiome project
-
A consortium project with the goal of comprehensively describing the commensal microorganisms associated with health and disease in humans.
Rights and permissions
About this article
Cite this article
Levy, M., Kolodziejczyk, A., Thaiss, C. et al. Dysbiosis and the immune system. Nat Rev Immunol 17, 219–232 (2017). https://doi.org/10.1038/nri.2017.7
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2017.7
This article is cited by
-
Cross-reactive CD8+ T cell responses to tumor-associated antigens (TAAs) and homologous microbiota-derived antigens (MoAs)
Journal of Experimental & Clinical Cancer Research (2024)
-
Gut dysbiosis induces the development of depression-like behavior through abnormal synapse pruning in microglia-mediated by complement C3
Microbiome (2024)
-
Fermented foods and gastrointestinal health: underlying mechanisms
Nature Reviews Gastroenterology & Hepatology (2024)
-
Leaf microbiome dysbiosis triggered by T2SS-dependent enzyme secretion from opportunistic Xanthomonas pathogens
Nature Microbiology (2024)
-
Dermal injury drives a skin to gut axis that disrupts the intestinal microbiome and intestinal immune homeostasis in mice
Nature Communications (2024)