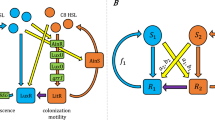
Key Points
-
Quorum sensing is a cell–cell communication process that enables bacteria to obtain information about cell density and species composition of the vicinal community and adjust their gene expression profiles accordingly. Quorum sensing involves the production, release and detection of extracellular signalling molecules known as autoinducers. Group-wide detection of autoinducers enables bacteria to collectively execute behaviours.
-
Autoinducers are small molecules that control quorum sensing. In Gram-negative bacteria, autoinducers are often produced from S-adenosylmethionine (SAM). Autoinducers interact with specific receptors to elicit behaviours that are controlled by quorum sensing.
-
Quorum sensing receptors are either membrane-bound histidine sensor kinases or cytoplasmic transcription factors.
-
Autoinduction occurs when the detection of autoinducers induces the increased production of the same autoinducer molecule, forming a feed-forward regulatory loop. Other features, such as positive and negative feedback loops and small regulatory RNAs, optimize the integration of the autoinducer-encoded information and provide ideal quorum sensing dynamics.
-
Signal integration is a process that takes place in most Gram-negative bacteria when several autoinducers and receptors work in parallel, or in series, to synchronize functions that are controlled by quorum sensing. Processes such as bioluminescence, the production of virulence factors and the formation of biofilms are controlled by quorum sensing.
-
Quorum sensing shapes the composition of microbial communities. For example, bacterial species in the human gut microbiota produce and respond to autoinducers. There is increasing evidence that quorum sensing controls key physiological processes in the gut and may affect the virulence programmes of invading pathogens. Host cells are also known to produce autoinducer mimics.
-
Synthetic quorum sensing modulators are molecules that agonize or antagonize quorum sensing and they are being developed as anti-virulence medicines. Distinct from traditional antibiotics, quorum sensing modulators do not affect the growth of pathogenic bacteria, but rather, disrupt their virulence programmes.
Abstract
Bacteria use quorum sensing to orchestrate gene expression programmes that underlie collective behaviours. Quorum sensing relies on the production, release, detection and group-level response to extracellular signalling molecules, which are called autoinducers. Recent work has discovered new autoinducers in Gram-negative bacteria, shown how these molecules are recognized by cognate receptors, revealed new regulatory components that are embedded in canonical signalling circuits and identified novel regulatory network designs. In this Review we examine how, together, these features of quorum sensing signal–response systems combine to control collective behaviours in Gram-negative bacteria and we discuss the implications for host–microbial associations and antibacterial therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bassler, B. L. & Losick, R. Bacterially speaking. Cell 125, 237–246 (2006).
LaSarre, B. & Federle, M. J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111 (2013).
Rutherford, S. T. & Bassler, B. L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 (2012).
Novick, R. P. & Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008).
Ng, W. L. & Bassler, B. L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222 (2009).
Papenfort, K. & Vogel, J. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8, 116–127 (2010).
Schuster, M., Sexton, D. J., Diggle, S. P. & Greenberg, E. P. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63 (2013).
Grote, J., Krysciak, D. & Streit, W. R. Phenotypic heterogeneity, a phenomenon that may explain why quorum sensing does not always result in truly homogenous cell behavior. Appl. Environ. Microbiol. 81, 5280–5289 (2015).
Carcamo-Oyarce, G., Lumjiaktase, P., Kummerli, R. & Eberl, L. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat. Commun. 6, 5945 (2015). This paper reports that information that is contained in autoinducers can regulate the behaviour of individual P. putida cells to promote heterogeneity.
Vogt, S. L., Pena-Diaz, J. & Finlay, B. B. Chemical communication in the gut: effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe 34, 106–115 (2015).
Gill, E. E., Franco, O. L. & Hancock, R. E. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 85, 56–78 (2015).
Ke, X., Miller, L. C. & Bassler, B. L. Determinants governing ligand specificity of the Vibrio harveyi LuxN quorum-sensing receptor. Mol. Microbiol. 95, 127–142 (2015).
Xavier, K. B. & Bassler, B. L. Interference with AI-2-mediated bacterial cell–cell communication. Nature 437, 750–753 (2005).
Galloway, W. R., Hodgkinson, J. T., Bowden, S. D., Welch, M. & Spring, D. R. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67 (2011).
Case, R. J., Labbate, M. & Kjelleberg, S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349 (2008).
von Bodman, S. B., Willey, J. M. & Diggle, S. P. Cell–cell communication in bacteria: united we stand. J. Bacteriol. 190, 4377–4391 (2008).
Schaefer, A. L. et al. A new class of homoserine lactone quorum-sensing signals. Nature 454, 595–599 (2008). This study identifies a new type of autoinducer, p -coumaroyl-HSL, from R. palustris.
Lindemann, A. et al. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc. Natl Acad. Sci. USA 108, 16765–16770 (2011).
Ahlgren, N. A., Harwood, C. S., Schaefer, A. L., Giraud, E. & Greenberg, E. P. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc. Natl Acad. Sci. USA 108, 7183–7188 (2011).
Flavier, A. B., Clough, S. J., Schell, M. A. & Denny, T. P. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259 (1997).
Kai, K. et al. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. Chembiochem 16, 2309–2318 (2015).
Genin, S. & Denny, T. P. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 (2012).
Tao, F., Swarup, S. & Zhang, L. H. Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environ. Microbiol. 12, 3159–3170 (2010).
Ryan, R. P., An, S. Q., Allan, J. H., McCarthy, Y. & Dow, J. M. The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 11, e1004986 (2015).
Zhou, L. et al. The multiple DSF-family QS signals are synthesized from carbohydrate and branched-chain amino acids via the FAS elongation cycle. Sci. Rep. 5, 13294 (2015).
Deng, Y., Wu, J., Eberl, L. & Zhang, L. H. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl. Environ. Microbiol. 76, 4675–4683 (2010).
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K. & Bassler, B. L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314 (2002).
van Kessel, J. C., Rutherford, S. T., Shao, Y., Utria, A. F. & Bassler, B. L. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J. Bacteriol. 195, 436–443 (2013).
Bassler, B. L., Wright, M., Showalter, R. E. & Silverman, M. R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9, 773–786 (1993).
Cao, J. G. & Meighen, E. A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264, 21670–21676 (1989).
Hanzelka, B. L. et al. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J. Bacteriol. 181, 5766–5770 (1999).
Higgins, D. A. et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886 (2007).
Kelly, R. C. et al. The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA. Nat. Chem. Biol. 5, 891–895 (2009).
Ng, W. L. et al. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 79, 1407–1417 (2011).
Wei, Y., Perez, L. J., Ng, W. L., Semmelhack, M. F. & Bassler, B. L. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem. Biol. 6, 356–365 (2011).
Spirig, T. et al. The Legionella autoinducer synthase LqsA produces an alpha-hydroxyketone signaling molecule. J. Biol. Chem. 283, 18113–18123 (2008).
Hornung, C. et al. The Janthinobacterium sp. HH01 genome encodes a homologue of the V. cholerae CqsA and L. pneumophila LqsA autoinducer synthases. PLoS ONE 8, e55045 (2013).
Simon, S. et al. Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1–Cdc42–ARHGEF9-dependent pathway. PLoS Pathog. 11, e1005307 (2015).
Schauder, S., Shokat, K., Surette, M. G. & Bassler, B. L. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476 (2001).
Pereira, C. S., Thompson, J. A. & Xavier, K. B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181 (2013).
Chen, X. et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549 (2002).
Miller, S. T. et al. Salmonella Typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15, 677–687 (2004).
Surette, M. G., Miller, M. B. & Bassler, B. L. Quorum sensing in Escherichia coli. Salmonella Typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl Acad. Sci. USA 96, 1639–1644 (1999).
Duan, K., Dammel, C., Stein, J., Rabin, H. & Surette, M. G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491 (2003).
Campbell, J., Lin, Q., Geske, G. D. & Blackwell, H. E. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 4, 1051–1059 (2009).
Lee, J. et al. A cell–cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 9, 339–343 (2013).
Pesci, E. C. et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 96, 11229–11234 (1999).
Lee, J. & Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41 (2015).
Heeb, S. et al. Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 35, 247–274 (2011).
Brameyer, S., Kresovic, D., Bode, H. B. & Heermann, R. Dialkylresorcinols as bacterial signaling molecules. Proc. Natl Acad. Sci. USA 112, 572–577 (2015).
Brachmann, A. O. et al. Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 9, 573–578 (2013). This paper reports the identification of a new class of pyrone autoinducer molecules in P. luminescens.
Fuchs, S. W. et al. Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occurring ketosynthase. Angew. Chem. Int. Ed Engl. 52, 4108–4112 (2013).
Smith, D. et al. Variations on a theme: diverse N-acyl homoserine lactone-mediated quorum sensing mechanisms in Gram-negative bacteria. Sci. Prog. 89, 167–211 (2006).
Swem, L. R. et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35, 143–153 (2009).
Zhang, R. G. et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417, 971–974 (2002). This paper reports the first crystal structure of a LuxR-type quorum sensing receptor, in this case, in complex with its cognate autoinducer and promoter DNA.
Engebrecht, J., Nealson, K. & Silverman, M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781 (1983).
Engebrecht, J. & Silverman, M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl Acad. Sci. USA 81, 4154–4158 (1984).
Stevens, A. M., Dolan, K. M. & Greenberg, E. P. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl Acad. Sci. USA 91, 12619–12623 (1994).
Schu, D. J., Scruggs, J. M., Geissinger, J. S., Michel, K. G. & Stevens, A. M. Acyl-homoserine lactone recognition and response hindering the quorum-sensing regulator EsaR. PLoS ONE 9, e107687 (2014).
von Bodman, S. B., Majerczak, D. R. & Coplin, D. L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl Acad. Sci. USA 95, 7687–7692 (1998).
Vannini, A. et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21, 4393–4401 (2002).
Chen, G., Jeffrey, P. D., Fuqua, C., Shi, Y. & Chen, L. Structural basis for antiactivation in bacterial quorum sensing. Proc. Natl Acad. Sci. USA 104, 16474–16479 (2007).
Lintz, M. J., Oinuma, K., Wysoczynski, C. L., Greenberg, E. P. & Churchill, M. E. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl Acad. Sci. USA 108, 15763–15768 (2011).
Chen, G. et al. A strategy for antagonizing quorum sensing. Mol. Cell 42, 199–209 (2011).
Bottomley, M. J., Muraglia, E., Bazzo, R. & Carfi, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 282, 13592–13600 (2007).
Yao, Y. et al. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol. 355, 262–273 (2006).
Galperin, M. Y., Nikolskaya, A. N. & Koonin, E. V. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203, 11–21 (2001).
Galperin, M. Y. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 (2006).
Li, Z. & Nair, S. K. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 21, 1403–1417 (2012).
Hudaiberdiev, S. et al. Census of solo LuxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol. 5, 20 (2015).
Lee, J. H., Lequette, Y. & Greenberg, E. P. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59, 602–609 (2006).
Riedel, K. et al. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262 (2001).
Swem, L. R., Swem, D. L., Wingreen, N. S. & Bassler, B. L. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134, 461–473 (2008).
Neiditch, M. B., Federle, M. J., Miller, S. T., Bassler, B. L. & Hughson, F. M. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18, 507–518 (2005).
Neiditch, M. B. et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126, 1095–1108 (2006).
Ng, W. L. et al. Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc. Natl Acad. Sci. USA 107, 5575–5580 (2010).
Pereira, C. S., de Regt, A. K., Brito, P. H., Miller, S. T. & Xavier, K. B. Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol. 191, 6975–6987 (2009).
Elias, S. & Banin, E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004 (2012).
Goryachev, A. B. Design principles of the bacterial quorum sensing gene networks. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 45–60 (2009).
Jimenez, P. N. et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65 (2012).
Seed, P. C., Passador, L. & Iglewski, B. H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177, 654–659 (1995).
Deziel, E. et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl Acad. Sci. USA 101, 1339–1344 (2004).
Winson, M. K. et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 92, 9427–9431 (1995).
Ventre, I. et al. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Mol. Microbiol. 48, 187–198 (2003).
McKnight, S. L., Iglewski, B. H. & Pesci, E. C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182, 2702–2708 (2000).
Cao, H. et al. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl Acad. Sci. USA 98, 14613–14618 (2001).
Hoffman, L. R. et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8, 66–70 (2009).
Zhu, J. & Winans, S. C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl Acad. Sci. USA 98, 1507–1512 (2001).
Freeman, J. A. & Bassler, B. L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31, 665–677 (1999).
Lilley, B. N. & Bassler, B. L. Regulation of quorum sensing in Vibrio harveyi by LuxO and σ54. Mol. Microbiol. 36, 940–954 (2000).
Lenz, D. H. et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82 (2004).
Vogel, J. & Luisi, B. F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011).
Shao, Y., Feng, L., Rutherford, S. T., Papenfort, K. & Bassler, B. L. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 32, 2158–2171 (2013).
Wei, Y., Ng, W. L., Cong, J. & Bassler, B. L. Ligand and antagonist driven regulation of the Vibrio cholerae quorum-sensing receptor CqsS. Mol. Microbiol. 83, 1095–1108 (2012).
Teng, S. W. et al. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol. Syst. Biol. 7, 491 (2011).
Tu, K. C., Long, T., Svenningsen, S. L., Wingreen, N. S. & Bassler, B. L. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell 37, 567–579 (2010).
Feng, L. et al. A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell 160, 228–240 (2015). This study combines experimental and theoretical work to determine the molecular mechanisms that underlie the regulation of target mRNAs by the Qrr small regulatory RNAs.
Sourjik, V. & Wingreen, N. S. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268 (2012).
Svenningsen, S. L., Tu, K. C. & Bassler, B. L. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 28, 429–439 (2009).
Hunter, G. A., Vasquez, F. G. & Keener, J. P. A mathematical model and quantitative comparison of the small RNA circuit in the Vibrio harveyi and Vibrio cholerae quorum sensing systems. Phys. Biol. 10, 046007 (2013).
Wang, Y., Tu, K. C., Ong, N. P., Bassler, B. L. & Wingreen, N. S. Protein-level fluctuation correlation at the microcolony level and its application to the Vibrio harveyi quorum-sensing circuit. Biophys. J. 100, 3045–3053 (2011).
Svenningsen, S. L., Waters, C. M. & Bassler, B. L. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 22, 226–238 (2008).
Rutherford, S. T., van Kessel, J. C., Shao, Y. & Bassler, B. L. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408 (2011).
Lin, W., Kovacikova, G. & Skorupski, K. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187, 3013–3019 (2005).
Long, T. et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 7, e68 (2009).
Nadell, C. D., Xavier, J. B., Levin, S. A. & Foster, K. R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 6, e14 (2008).
Fuqua, W. C., Winans, S. C. & Greenberg, E. P. Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994).
Bassler, B. L. Small talk: cell-to-cell communication in bacteria. Cell 109, 421–424 (2002).
Pacheco, A. R. & Sperandio, V. Inter-kingdom signaling: chemical language between bacteria and host. Curr. Opin. Microbiol. 12, 192–198 (2009).
Srivastava, D. & Waters, C. M. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194, 4485–4493 (2012).
Pesavento, C. & Hengge, R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 12, 170–176 (2009).
Deng, Y. et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl Acad. Sci. USA 109, 15479–15484 (2012).
Kalia, D. et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341 (2013).
Persat, A. et al. The mechanical world of bacteria. Cell 161, 988–997 (2015).
Drescher, K., Nadell, C. D., Stone, H. A., Wingreen, N. S. & Bassler, B. L. Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55 (2014).
Kim, M. K., Ingremeau, F., Zhao, A., Bassler, B. L. & Stone, H. A. Local and global consequences of flow on bacterial quorum sensing. Nat. Microbiol. 1, 15005 (2016). This paper shows that fluid flow and topography influence gene expression that is controlled by quorum sensing in V. cholerae and Staphylococcus aureus in non-intuitive ways.
Rickard, A. H., Campagna, S. R. & Kolenbrander, P. E. Autoinducer-2 is produced in saliva-fed flow conditions relevant to natural oral biofilms. J. Appl. Microbiol. 105, 2096–2103 (2008).
Shao, Y. & Bassler, B. L. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol. Microbiol. 92, 921–930 (2014).
Zheng, J., Shin, O. S., Cameron, D. E. & Mekalanos, J. J. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl Acad. Sci. USA 107, 21128–21133 (2010).
MacIntyre, D. L., Miyata, S. T., Kitaoka, M. & Pukatzki, S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl Acad. Sci. USA 107, 19520–19524 (2010).
Borgeaud, S., Metzger, L. C., Scrignari, T. & Blokesch, M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015). This report finds that V. cholerae uses type VI secretion to scavenge DNA from neighbouring cells.
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C. & Xavier, K. B. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871 (2015). This study shows that AI-2 quorum sensing signalling shapes the composition of the gut microbiota.
Hsiao, A. et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014). This study reveals a role for particular species in the gut microbiota in recovery from infection with V. cholerae through the LuxR transcription factor VqmA.
Liu, Z., Hsiao, A., Joelsson, A. & Zhu, J. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J. Bacteriol. 188, 2446–2453 (2006).
Papenfort, K., Forstner, K. U., Cong, J. P., Sharma, C. M. & Bassler, B. L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl Acad. Sci. USA 112, E766–E775 (2015).
Zargar, A. et al. Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. mBio 6, e00025 (2015).
Shiner, E. K. et al. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell. Microbiol. 8, 1601–1610 (2006).
Karavolos, M. H., Winzer, K., Williams, P. & Khan, C. M. Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465 (2013).
Ismail, A. S., Valastyan, J. S. & Bassler, B. L. A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480 (2016). This study finds that human epithelial cells produce an AI-2 mimic that is detected by the LuxPQ and LsrB quorum sensing receptors.
Helman, Y. & Chernin, L. Silencing the mob: disrupting quorum sensing as a means to fight plant disease. Mol. Plant Pathol. 16, 316–329 (2015).
Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015).
Plener, L. et al. The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J. Bacteriol. 197, 1747–1756 (2015).
Anetzberger, C., Pirch, T. & Jung, K. Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol. Microbiol. 73, 267–277 (2009).
Franks, N. R. et al. Not everything that counts can be counted: ants use multiple metrics for a single nest trait. Proc. Biol. Sci. 273, 165–169 (2006).
Seeley, T. D. & Visscher, P. K. Group decision making in nest-site selection by honey bees. Apidologie 35, 101–116 (2004).
Chen, C. C. et al. Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 161, 277–290 (2015). This paper reports that quorum sensing principles are applicable in eukaryotic systems.
McInnis, C. E. & Blackwell, H. E. Design, synthesis, and biological evaluation of abiotic, non-lactone modulators of LuxR-type quorum sensing. Bioorg. Med. Chem. 19, 4812–4819 (2011).
Geske, G. D., Mattmann, M. E. & Blackwell, H. E. Evaluation of a focused library of N-aryl l-homoserine lactones reveals a new set of potent quorum sensing modulators. Bioorg. Med. Chem. Lett. 18, 5978–5981 (2008).
Stacy, D. M. et al. Synthesis and biological evaluation of triazole-containing N-acyl homoserine lactones as quorum sensing modulators. Org. Biomol. Chem. 11, 938–954 (2013).
O'Loughlin, C. T. et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl Acad. Sci. USA 110, 17981–17986 (2013). This study identifies the quorum sensing inhibitor meta-bromo-thiolactone (mBTL), and shows that it blocks the production of virulence factors and the formation of biofilms in P. aeruginosa.
Ishida, T. et al. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 73, 3183–3188 (2007).
Welsh, M. A., Eibergen, N. R., Moore, J. D. & Blackwell, H. E. Small molecule disruption of quorum sensing cross-regulation in Pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J. Am. Chem. Soc. 137, 1510–1519 (2015).
Welsh, M. A. & Blackwell, H. E. Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 23, 361–369 (2016).
Ilangovan, A. et al. Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog. 9, e1003508 (2013).
Lu, C. et al. Discovery of antagonists of PqsR, a key player in 2-alkyl-4-quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 19, 381–390 (2012).
Lu, C., Maurer, C. K., Kirsch, B., Steinbach, A. & Hartmann, R. W. Overcoming the unexpected functional inversion of a PqsR antagonist in Pseudomonas aeruginosa: an in vivo potent antivirulence agent targeting pqs quorum sensing. Angew. Chem. Int. Ed Engl. 53, 1109–1112 (2014).
Starkey, M. et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 10, e1004321 (2014).
Calfee, M. W., Coleman, J. P. & Pesci, E. C. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 98, 11633–11637 (2001).
Chang, C. Y. et al. Non-antibiotic quorum sensing inhibitors acting against N-acyl homoserine lactone synthase as druggable target. Sci. Rep. 4, 7245 (2014).
Bolitho, M. E. et al. Small molecule probes of the receptor binding site in the Vibrio cholerae CAI-1 quorum sensing circuit. Bioorg. Med. Chem. 19, 6906–6918 (2011).
Faloon, P. et al. in Probe Reports from the NIH Molecular Libraries Program (Bethesda (MD), 2010).
Ng, W. L., Perez, L., Cong, J., Semmelhack, M. F. & Bassler, B. L. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 8, e1002767 (2012).
Gutierrez, J. A. et al. Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 5, 251–257 (2009).
Silva, A. J., Parker, W. B., Allan, P. W., Ayala, J. C. & Benitez, J. A. Role of methylthioadenosine/S- adenosylhomocysteine nucleosidase in Vibrio cholerae cellular communication and biofilm development. Biochem. Biophys. Res. Commun. 461, 65–69 (2015).
Kohler, T., Perron, G. G., Buckling, A. & van Delden, C. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 6, e1000883 (2010).
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, the US National Institutes of Health (NIH; grant 5R01GM065859) and the US National Science Foundation (grant MCB-0948112 to B.L.B.). K.P. was supported by the Deutsche Forschungsgemeinschaft (DFG; grant PA2820/1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Public goods
-
Common-pool resources that are frequently present in biological and social systems. Public goods are available to all members of the community, irrespective of whether a member contributed to their production. Therefore, public goods are prone to exploitation by non-producers.
- Two-component systems
-
A large group of signal-transduction circuits that typically consist of a membrane-bound histidine sensor kinase that detects a specific environmental stimulus and a cognate response regulator that mediates the cellular response, primarily through transcriptional regulation of target genes.
- Feed-forward loop
-
A common regulatory network motif in biological pathways. The feed-forward loop is composed of two input factors (usually transcriptional regulators), one of which regulates the other, such that both factors jointly regulate downstream target genes.
- Small RNAs
-
(sRNAs). Bacterial sRNAs are a heterogeneous group of post-transcriptional regulators that often act in conjunction with the chaperone Hfq.
- Bet-hedging
-
A survival strategy that reduces the temporal variance in fitness at the expense of reduced arithmetic mean fitness.
- GAF and PAS domains
-
Domains that are often conserved in signalling proteins in which they function as ligand-binding domains.
- van der Waals interactions
-
Weak attractive or repulsive forces between molecules or atomic groups that do not result from covalent bonds or electrostatic interactions between ions or ionic groups.
- ATP-binding cassette transporter
-
(ABC transporter). A member of a large superfamily of small molecule transport systems that are present in all phyla.
- σ54
-
An alternative sigma factor in bacteria that is encoded by the rpoN gene, which was originally identified as a regulator of genes that are involved in nitrogen metabolism.
- Hfq
-
A globally acting RNA-binding protein that facilitates the base pairing of bacterial small RNAs with their target mRNAs.
- Cyclic dimeric guanosine monophosphate
-
(c-di-GMP). A second-messenger molecule that is used in signal transduction in various bacteria.
- Cyclic adenosine monophosphate
-
(cAMP). A second-messenger molecule that is important in many biological processes in organisms, ranging from bacteria to humans.
- GGDEF and EAL domains
-
Protein domains that are ubiquitous in bacteria and function to synthesize and degrade the intracellular signalling molecule cyclic dimeric guanosine monophosphate (c-di-GMP), respectively.
- Type VI secretion
-
Systems that are used by Gram-negative bacteria to inject effector proteins and virulence factors from across the interior of one bacterial cell into another cell called the prey.
- Horizontal gene transfer
-
The exchange of genetic information between organisms in a manner other than by traditional reproduction. Horizontal gene transfer is key for the acquisition of antibiotic resistance in bacteria and also has an important role in evolution and the generation of diversity.
- Persister cells
-
Isogenic members of a bacterial population that have entered a non-growing or extremely slow-growing physiological state, which makes them tolerant to a wide range of antimicrobials.
Rights and permissions
About this article
Cite this article
Papenfort, K., Bassler, B. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 14, 576–588 (2016). https://doi.org/10.1038/nrmicro.2016.89
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.89
This article is cited by
-
Methanogenic partner influences cell aggregation and signalling of Syntrophobacterium fumaroxidans
Applied Microbiology and Biotechnology (2024)
-
Enhancing the predictive capability of a mathematical model for pseudomonas aeruginosa through artificial neural networks
International Journal of Information Technology (2024)
-
D-histidine combated biofilm formation and enhanced the effect of amikacin against Pseudomonas aeruginosa in vitro
Archives of Microbiology (2024)
-
Comprehensive genome analysis of Burkholderia contaminans SK875, a quorum-sensing strain isolated from the swine
AMB Express (2023)
-
A narrative review on bacterial biofilm: its formation, clinical aspects and inhibition strategies
Future Journal of Pharmaceutical Sciences (2023)