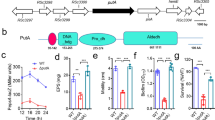
Abstract
Ethanolamine is a compound that can be readily derived from cell membranes and that some bacteria can use as a source of carbon and/or nitrogen. The complex biology and chemistry of this process has been under investigation since the 1970s, primarily in one or two species. However, recent investigations into ethanolamine utilization have revealed important and intriguing differences in gene content and regulatory mechanisms among the bacteria that harbour this catabolic ability. In addition, many reports have connected this process to bacterial pathogenesis. In this Progress article, I discuss the latest research on the phylogeny and regulation of ethanolamine utilization and its possible roles in bacterial pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Randle, C. L., Albro, P. W. & Dittmer, J. C. The phosphoglyceride composition of Gram-negative bacteria and the changes in composition during growth. Biochim. Biophys. Acta 187, 214–220 (1969).
White, D. A. in Form and Function of Phospholipids (eds Ansell, G. B., Hawthorne, J. N. & Dawson, R. M. C.) 441–482 (Elsevier, New York, 1973).
Larson, T. J., Ehrmann, M. & Boos, W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 258, 5428–5432 (1983).
Proulx, P. & Fung, C. K. Metabolism of phosphoglycerides in E. coli. IV. The positional specificity and properties of phospholipase A. Can. J. Biochem. 47, 1125–1128 (1969).
Roof, D. M. & Roth, J. R. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170, 3855–3863 (1988).
Chang, G. W. & Chang, J. T. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254, 150–151 (1975).
Del Papa, M. F. & Perego, M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190, 7147–7156 (2008).
Blackwell, C. M., Scarlett, F. A. & Turner, J. M. Ethanolamine catabolism by bacteria, including Escherichia coli. Biochem. Soc. Trans. 4, 495–497 (1976).
Bradbeer, C. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J. Biol. Chem. 240, 4675–4681 (1965).
Bradbeer, C. The clostridial fermentations of choline and ethanolamine. 1. Preparation and properties of cell-free extracts. J. Biol. Chem. 240, 4669–4674 (1965).
Jones, P. W. & Turner, J. M. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J. Gen. Microbiol. 130, 299–308 (1984).
Fox, K. A. et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl Acad. Sci. USA 106, 4435–4440 (2009).
Tsoy, O., Ravcheev, D. & Mushegian, A. Comparative genomics of ethanolamine utilization. J. Bacteriol. 191, 7157–7164 (2009).
Roof, D. M. & Roth, J. R. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171, 3316–3323 (1989).
Sheppard, D. E., Penrod, J. T., Bobik, T., Kofoid, E. & Roth, J. R. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186, 7635–7644 (2004).
Stojiljkovic, I., Baumler, A. J. & Heffron, F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177, 1357–1366 (1995).
Kofoid, E., Rappleye, C., Stojiljkovic, I. & Roth, J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181, 5317–5329 (1999).
Yeates, T. O., Kerfeld, C. A., Heinhorst, S., Cannon, G. C. & Shively, J. M. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nature Rev. Microbiol. 6, 681–691 (2008).
Brinsmade, S. R., Paldon, T. & Escalante-Semerena, J. C. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187, 8039–8046 (2005).
Penrod, J. T. & Roth, J. R. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188, 2865–2874 (2006).
Scarlett, F. A. & Turner, J. M. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia lyase in Escherichia coli and Klebsiella aerogenes. J. Gen. Microbiol. 95, 173–176 (1976).
Buan, N. R., Suh, S. J. & Escalante-Semerena, J. C. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J. Bacteriol. 186, 5708–5714 (2004).
Brinsmade, S. R. & Escalante-Semerena, J. C. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186, 1890–1892 (2004).
Starai, V. J., Garrity, J. & Escalante-Semerena, J. C. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology 151, 3793–3801 (2005).
Mori, K., Bando, R., Hieda, N. & Toraya, T. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J. Bacteriol. 186, 6845–6854 (2004).
Penrod, J. T., Mace, C. C. & Roth, J. R. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J. Bacteriol. 186, 6885–6890 (2004).
Roof, D. M. & Roth, J. R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174, 6634–6643 (1992).
Blackwell, C. M. & Turner, J. M. Microbial metabolism of amino alcohols. Formation of coenzyme B12-dependent ethanolamine ammonia-lyase and its concerted induction in Escherichia coli. Biochem. J. 176, 751–757 (1978).
Sheppard, D. E. & Roth, J. R. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J. Bacteriol. 176, 1287–1296 (1994).
Laub, M. T. & Goulian, M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41, 121–145 (2007).
Galperin, M. Y. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 (2006).
Shu, C. J. & Zhulin, I. B. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27, 3–5 (2002).
Chai, W. & Stewart, V. NasR, a novel RNA-binding protein, mediates nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J. Mol. Biol. 283, 339–351 (1998).
Chai, W. & Stewart, V. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J. Mol. Biol. 292, 203–216 (1999).
Wilson, S. A., Wachira, S. J., Norman, R. A., Pearl, L. H. & Drew, R. E. Transcription antitermination regulation of the Pseudomonas aeruginosa amidase operon. EMBO J. 15, 5907–5916 (1996).
Barrick, J. E. & Breaker, R. R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8, R239 (2007).
Winkler, W. C. & Breaker, R. R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59, 487–517 (2005).
Morth, J. P., Feng, V., Perry, L. J., Svergun, D. I. & Tucker, P. A. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure 12, 1595–1605 (2004).
O'Hara, B. P. et al. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 18, 5175–5186 (1999).
Cotton, P. B. Non-dietary lipid in the intestinal lumen. Gut 13, 675–681 (1972).
Toledo-Arana, A. et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956 (2009).
Berka, R. M. & Vasil, M. L. Phospholipase C (heat-labile hemolysin) of Pseudomonas aeruginosa: purification and preliminary characterization. J. Bacteriol. 152, 239–245 (1982).
Lema, G., Dryja, D., Vargas, I. & Enhorning, G. Pseudomonas aeruginosa from patients with cystic fibrosis affects function of pulmonary surfactant. Pediatr. Res. 47, 121–126 (2000).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Lawhon, S. D. et al. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48, 1633–1645 (2003).
Kelly, A. et al. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150, 2037–2053 (2004).
Bourgogne, A., Hilsenbeck, S. G., Dunny, G. M. & Murray, B. E. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188, 2875–2884 (2006).
Garsin, D. A. et al. A simple model host for identifying Gram-positive virulence factors. Proc. Natl Acad. Sci. USA 98, 10892–10897 (2001).
Maadani, A., Fox, K. A., Mylonakis, E. & Garsin, D. A. Enterococcus faecalis mutations affecting virulence in the C. elegans model host. Infect. Immun. 75, 2634–2637 (2007).
Joseph, B. et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568 (2006).
Munch, A., Stingl, L., Jung, K. & Heermann, R. Photorhabdus luminescens genes induced upon insect infection. BMC Genomics 9, 229 (2008).
Yang, S. et al. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol. Plant Microbe Interact. 17, 999–1008 (2004).
Forouhar, F. et al. Functional insights from structural genomics. J. Struct. Funct. Genomics 8, 37–44 (2007).
Sagermann, M., Ohtaki, A. & Nikolakakis, K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl Acad. Sci. USA 106, 8883–8887 (2009).
Tanaka, S., Sawaya, M. R. & Yeates, T. O. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327, 81–84 (2010).
Acknowledgements
I thank W. C. Winkler and K. A. Fox for critical reading of the manuscript. This work was funded by the US National Institute of Allergy and Infectious Diseases (award R21AI078104).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Salmonella enterica subsp. enterica serovar Typhimurium
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Garsin, D. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8, 290–295 (2010). https://doi.org/10.1038/nrmicro2334
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2334
This article is cited by
-
Turnover of strain-level diversity modulates functional traits in the honeybee gut microbiome between nurses and foragers
Genome Biology (2023)
-
Whole-Genome Sequence and Pathogenicity Analysis of Providencia Heimbachae Causing Diarrhea in Weaned Piglets
Current Microbiology (2023)
-
Design and validation of a dual-fluorescence reporter system to monitor bacterial gene expression in the gut environment
Applied Microbiology and Biotechnology (2023)
-
A Systemic Review on Fitness and Survival of Salmonella in Dynamic Environment and Conceivable Ways of Its Mitigation
Indian Journal of Microbiology (2023)
-
Comparative genomics reveals low levels of inter- and intraspecies diversity in the causal agents of dwarf and common bunt of wheat and hint at conspecificity of Tilletia caries and T. laevis
IMA Fungus (2022)