Abstract
Nucleosome assembly following DNA replication, DNA repair and gene transcription is critical for the maintenance of genome stability and epigenetic information. Nucleosomes are assembled by replication-coupled or replication-independent pathways with the aid of histone chaperone proteins. How these different nucleosome assembly pathways are regulated remains relatively unclear. Recent studies have provided insight into the mechanisms and the roles of histone chaperones in regulating nucleosome assembly. Alterations or mutations in factors involved in nucleosome assembly have also been implicated in cancer and other human diseases. This review highlights the recent progress and outlines future challenges in the field.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ransom, M., Dennehey, B.K. & Tyler, J.K. Chaperoning histones during DNA replication and repair. Cell 140, 183–195 (2010).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Moazed, D. Mechanisms for the inheritance of chromatin states. Cell 146, 510–518 (2011).
Margueron, R. & Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Smith, S. & Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991).
Torigoe, S.E., Urwin, D.L., Ishii, H., Smith, D.E. & Kadonaga, J.T. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol. Cell 43, 638–648 (2011).
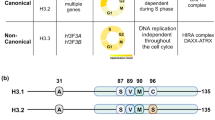
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004).Shows that histone H3.1 and H3.3 are deposited by distinct histone chaperones in replication- coupled and replication-independent processes, respectively.
Drané, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010).
Goldberg, A.D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).Refs. 9,10 identify Daxx-ATRX as an H3.3–H4 histone chaperone, and data in ref. 10 indicate that Daxx and HIRA deposit H3.3 at different chromatin regions.
McKnight, S.L. & Miller, O.L. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophilia melanogaster embryo. Cell 12, 795–804 (1977).
Stillman, B. Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565 (1986).
Smith, D.J. & Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012).
Tyler, J.K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999).
Han, J. et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315, 653–655 (2007).
Hoek, M. & Stillman, B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100, 12183–12188 (2003).
Xu, M. et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 (2010).
Campos, E.I. et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 (2010). Suggests that predeposition histone H3.1–H4 associates with multiple chaperones, aiding in histone synthesis or stability, modification and nuclear import.
Cook, A.J., Gurard-Levin, Z.A., Vassias, I. & Almouzni, G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3–H4 in the histone supply chain. Mol. Cell 44, 918–927 (2011).
English, C.M., Adkins, M.W., Carson, J.J., Churchill, M.E. & Tyler, J.K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).Structural and functional analyses of Asf1–H3–H4 reveal that Asf1 forms a complex with the H3–H4 heterodimer through the H3 interface involved in the formation of a (H3–H4) 2 tetramer.
English, C.M., Maluf, N.K., Tripet, B., Churchill, M.E. & Tyler, J.K. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry 44, 13673–13682 (2005).
Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M.M. & Wu, C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 (2007).
Dunleavy, E.M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009).
Foltz, D.R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009).
Hu, H. et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 (2011).
Zhou, Z. et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237 (2011). Refs. 25,26 demonstrate that HJURP and its yeast counterpart Scm3 bind to the dimeric form of CenH3–H4 and prevent the spontaneous association of CenH3–H4 with DNA.
Jasencakova, Z. et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 (2010).
Driscoll, R., Hudson, A. & Jackson, S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 (2007).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Li, Q. et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 (2008).
Tyler, J.K. et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21, 6574–6584 (2001).
Krawitz, D.C., Kama, T. & Kaufman, P.D. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22, 614–625 (2002).
Donham, D.C. II., Scorgie, J.K. & Churchill, M.E. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 39, 5449–5458 (2011).
Winkler, D.D., Zhou, H., Dar, M.A., Zhang, Z. & Luger, K. Yeast CAF-1 assembles histone (H3–H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 40, 10139–10149 (2012).
Su, D. et al. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature 483, 104–107 (2012). Shows that Rtt106 binds (H3–H4) 2 tetramers and contains two histone-binding domains, an N-terminal oligomerization domain and tandem PH domains, which recognize H3 acetylated at lysine 56.
Zhang, W. et al. Structural plasticity of histones H3–H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat. Struct. Mol. Biol. 19, aaa–bbb (2012).
Fazly, A. et al. Histone chaperone Rtt106 promotes nucleosome formation using (H3–H4)2 tetramers. J. Biol. Chem. 287, 10753–10760 (2012).
Zunder, R.M., Antczak, A.J., Berger, J.M. & Rine, J. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc. Natl. Acad. Sci. USA 109, E144–E153 (2012).
Liu, Y. et al. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J. Biol. Chem. 285, 4251–4262 (2010).
Quivy, J.P., Grandi, P. & Almouzni, G. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 20, 2015–2027 (2001).
Nakano, S., Stillman, B. & Horvitz, H.R. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell 147, 1525–1536 (2011).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Sobel, R.E., Cook, R.G., Perry, C.A., Annunziato, A.T. & Allis, C.D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92, 1237–1241 (1995).
Alvarez, F. et al. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 (2011).
Kang, B. et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 25, 1359–1364 (2011). Shows that phosphorylation of histone H4 Ser47 by Pak2 differentially regulates H3.1–H4 and H3.3–H4 deposition by promoting the association of HIRA with H3.3–H4 and inhibiting the association of CAF-1 with H3.1–H4.
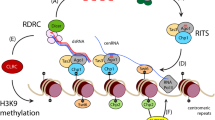
Loyola, A. et al. The HP1α-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10, 769–775 (2009).
Pinheiro, I. et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 (2012).
Parthun, M.R., Widom, J. & Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 (1996).
Ejlassi-Lassallette, A., Mocquard, E., Arnaud, M.C. & Thiriet, C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell 22, 245–255 (2011).
Zhang, H., Han, J., Kang, B., Burgess, R. & Zhang, Z. Human histone acetyltransferase HAT1 preferentially acetylates H4 molecules in H3.1–H4 dimers over H3.3–H4 dimers. J. Biol. Chem. 287, 6573–6581 (2012).
Ye, J. et al. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell 18, 123–130 (2005).
Yang, X. et al. HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44, 39–50 (2011). Shows that HAT4 is a new histone acetyltransferase that may be involved in replication-coupled nucleosome assembly in human cells.
Burgess, R.J., Zhou, H., Han, J. & Zhang, Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell 37, 469–480 (2010).
Das, C., Lucia, M.S., Hansen, K.C. & Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999).
Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 (2000).
Moggs, J.G. et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20, 1206–1218 (2000).
Groth, A. et al. Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007).
Franco, A.A., Lam, W.M., Burgers, P.M. & Kaufman, P.D. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19, 1365–1375 (2005).
Schulz, L.L. & Tyler, J.K. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 20, 488–490 (2006).
Tan, B.C., Chien, C.T., Hirose, S. & Lee, S.C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 25, 3975–3985 (2006).
Wittmeyer, J., Joss, L. & Formosa, T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38, 8961–8971 (1999).
Deal, R.B., Henikoff, J.G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010).
Szenker, E., Ray-Gallet, D. & Almouzni, G. The double face of the histone variant H3.3. Cell Res. 21, 421–434 (2011).
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Kappes, F. et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 25, 673–678 (2011).
Sawatsubashi, S. et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 24, 159–170 (2010).
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).Describes a SNAP-based assay to monitor H3.1 and H3.3 deposition in real time and shows that HIRA binds RNA polymerase II and DNA, aiding in the coordination of HIRA-mediated H3.3–H4 deposition and gene transcription.
Law, M.J. et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143, 367–378 (2010).
Iwase, S. et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 18, 769–776 (2011).
Katan-Khaykovich, Y. & Struhl, K. Splitting of H3–H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc. Natl. Acad. Sci. USA 108, 1296–1301 (2011).
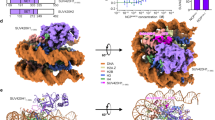
Liu, C.P. et al. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 19, 1287–1292 (2012).
Elsasser, S.J. et al. DAXX envelops an H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012). Refs. 72,73 show that Daxx binds an H3.3–H4 heterodimer and describe how the histone-binding domain of Daxx recognizes H3.3 preferentially over H3.1.
Rufiange, A., Jacques, P.E., Bhat, W., Robert, F. & Nourani, A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007).
Williams, S.K., Truong, D. & Tyler, J.K. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. USA 105, 9000–9005 (2008).
Guillemette, B. et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 7, e1001354 (2011).
Bokoch, G.M. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 (2003).
Zee, B.M., Levin, R.S., Dimaggio, P.A. & Garcia, B.A. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22 (2010).
Jamai, A., Imoberdorf, R.M. & Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007).
Selth, L. & Svejstrup, J.Q. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 282, 12358–12362 (2007).
Andrews, A.J., Downing, G., Brown, K., Park, Y.J. & Luger, K. A thermodynamic model for Nap1-histone interactions. J. Biol. Chem. 283, 32412–32418 (2008).
Mosammaparast, N., Ewart, C.S. & Pemberton, L.F. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538 (2002).
Ito, T., Bulger, M., Pazin, M.J., Kobayashi, R. & Kadonaga, J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997).
Andrews, A.J., Chen, X., Zevin, A., Stargell, L.A. & Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834–842 (2010).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Winkler, D.D., Muthurajan, U.M., Hieb, A.R. & Luger, K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 (2011).
Stuwe, T. et al. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. Proc. Natl. Acad. Sci. USA 105, 8884–8889 (2008).
VanDemark, A.P. et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22, 363–374 (2006).
Xin, H. et al. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol. Cell 35, 365–376 (2009).
Jamai, A., Puglisi, A. & Strubin, M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35, 377–383 (2009).
Batta, K., Zhang, Z., Yen, K., Goffman, D.B. & Pugh, B.F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 (2011).
Pavri, R. et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 (2006).
Yuan, J., Adamski, R. & Chen, J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 584, 3717–3724 (2010).
Heo, K. et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, 86–97 (2008).
Zhang, H., Roberts, D.N. & Cairns, B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231 (2005).
Jin, C. et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 (2009).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Luk, E. et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25, 357–368 (2007).
Straube, K., Blackwell, J.S. Jr. & Pemberton, L.F. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic 11, 185–197 (2010).
Costanzi, C. & Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Zhang, R. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005).
Iles, N., Rulten, S., El-Khamisy, S.F. & Caldecott, K.W. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27, 3793–3803 (2007).
Mehrotra, P.V. et al. DNA repair factor APLF is a histone chaperone. Mol. Cell 41, 46–55 (2011).
Lorain, S. et al. Structural organization of the WD repeat protein-encoding gene HIRA in the DiGeorge syndrome critical region of human chromosome 22. Genome Res. 6, 43–50 (1996).
Farrell, M.J. et al. HIRA, a DiGeorge syndrome candidate gene, is required for cardiac outflow tract septation. Circ. Res. 84, 127–135 (1999).
Wilming, L.G., Snoeren, C.A., van Rijswijk, A., Grosveld, F. & Meijers, C. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Hum. Mol. Genet. 6, 247–258 (1997).
Jiao, Y. et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 (2011).
Heaphy, C.M. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). Refs. 107–109 reveal mutations or alterations of the Daxx–ATRX–H3.3 deposition pathway in various cancers and describe the association of these mutations with alterations in telomeres and gene expression.
Wise-Draper, T.M. et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 69, 1792–1799 (2009).
Soekarman, D. et al. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood 79, 2990–2997 (1992).
Renella, R. et al. Codanin-1 mutations in congenital dyserythropoietic anemia type 1 affect HP1α localization in erythroblasts. Blood 117, 6928–6938 (2011).
Ask, K. et al. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 31, 2013–2023 (2012).
Corpet, A. et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30, 480–493 (2011).
Polo, S.E. et al. Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology 57, 716–724 (2010).
Verreault, A., Kaufman, P.D., Kobayashi, R. & Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 (1996).
Laskey, R.A., Honda, B.M., Mills, A.D. & Finch, J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420 (1978).
Han, J., Zhou, H., Li, Z., Xu, R.M. & Zhang, Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 282, 28587–28596 (2007).
Acknowledgements
We apologize to those whose work could not cited in this review, owing to space limitations. We thank G. Mer for use of the Rtt106–(H3–H4)2 complex structural model coordinates. This work is supported by grants to Z.Z. from the US National Institutes of Health (GM72719, GM81838, GM99722) and the National Science Foundation of China (NSFC) Collaboration grant (31210103914). Z.Z. is supported as a scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Burgess, R., Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20, 14–22 (2013). https://doi.org/10.1038/nsmb.2461
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2461
This article is cited by
-
Alzheimer’s Disease-Related Epigenetic Changes: Novel Therapeutic Targets
Molecular Neurobiology (2024)
-
Chromatin balances cell redox and energy homeostasis
Epigenetics & Chromatin (2023)
-
Reference compounds for characterizing cellular injury in high-content cellular morphology assays
Nature Communications (2023)
-
Structural comparisons reveal diverse binding modes between nucleosome assembly proteins and histones
Epigenetics & Chromatin (2022)
-
NPM2 in malignant peritoneal mesothelioma: from basic tumor biology to clinical medicine
World Journal of Surgical Oncology (2022)