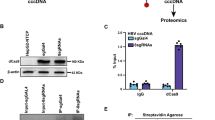
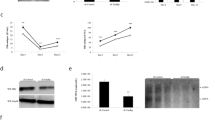
Abstract
Chronic hepatitis B virus (HBV) infections result in 887,000 deaths annually. The central challenge in curing HBV is eradication of the stable covalently closed circular DNA (cccDNA) form of the viral genome, which is formed by the repair of lesion-bearing HBV relaxed circular DNA delivered by the virions to hepatocytes. The complete and minimal set of host factors involved in cccDNA formation is unknown, largely due to the lack of a biochemical system that fully reconstitutes cccDNA formation. Here, we have developed experimental systems where various HBV relaxed-circular-DNA substrates are repaired to form cccDNA by both cell extracts and purified human proteins. Using yeast- and human-extract screenings, we identified five core components of lagging-strand synthesis as essential for cccDNA formation: proliferating cell nuclear antigen, the replication factor C complex, DNA polymerase δ, flap endonuclease 1 and DNA ligase 1. We reconstituted cccDNA formation with purified human homologues, establishing these as a minimal set of factors for cccDNA formation. We further demonstrated that treatment with the DNA-polymerase inhibitor aphidicolin diminishes cccDNA formation both in biochemical assays and in HBV-infected human cells. Together, our findings define key components in HBV cccDNA formation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Winer, B. Y. & Ploss, A. Determinants of hepatitis B and delta virus host tropism. Curr. Opin. Virol. 13, 109–116 (2015).
Nassal, M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984 (2015).
Yan, H. et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1, e00049 (2012).
Wang, G. H. & Seeger, C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71, 663–670 (1992).
Gerlich, W. H. & Robinson, W. S. Hepatitis B virus contains protein attached to the 5ʹ terminus of its complete DNA strand. Cell 21, 801–809 (1980).
Lucifora, J. & Protzer, U. Attacking hepatitis B virus cccDNA—the holy grail to hepatitis B cure. J. Hepatol. 64, S41–S48 (2016).
Koniger, C. et al. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl Acad. Sci. USA 111, E4244–E4253 (2014).
Gao, W. & Hu, J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 81, 6164–6174 (2007).
Guo, H. et al. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81, 12472–12484 (2007).
Hu, J. & Seeger, C. Hepadnavirus genome replication and persistence. Cold Spring Harb. Perspect. Med. 5, a021386 (2015).
Qi, Y. et al. DNA polymerase κ is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 12, e1005893 (2016).
Long, Q. et al. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 13, e1006784 (2017).
Cui, X. et al. Does tyrosyl DNA phosphodiesterase-2 play a role in hepatitis B virus genome repair? PLoS ONE 10, e0128401 (2015).
Kitamura, K. et al. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 14, e1007124 (2018).
Tang, L., Sheraz, M., McGrane, M., Chang, J. & Guo, J. T. DNA polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 15, e1007742 (2019).
Li, X., Zhao, J., Yuan, Q. & Xia, N. Detection of HBV covalently closed circular DNA. Viruses 9, 139 (2017).
Molnar-Kimber, K. L., Summers, J., Taylor, J. M. & Mason, W. S. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J. Virol. 45, 165–172 (1983).
Yan, Z. et al. HBVcircle: a novel tool to investigate hepatitis B virus covalently closed circular DNA. J. Hepatol. 66, 1149–1157 (2017).
Waga, S. & Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 (1998).
Winer, B. Y. et al. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat. Commun. 8, 125 (2017).
Wright, G. E., Hubscher, U., Khan, N. N., Focher, F. & Verri, A. Inhibitor analysis of calf thymus DNA polymerases α, δ and ϵ. FEBS Lett. 341, 128–130 (1994).
Qu, B., Ni, Y., Lempp, F. A., Vondran, F. W. R. & Urban, S. T5 exonuclease hydrolysis of hepatitis B virus replicative intermediates allows reliable quantification and fast drug efficacy testing of covalently closed circular DNA by PCR. J. Virol. 92, e01117-18 (2018).
Luo, J., Cui, X., Gao, L. & Hu, J. Identification of intermediate in hepatitis B virus CCC DNA formation and sensitive and selective CCC DNA detection. J. Virol. 91, e00539-17 (2017).
Cai, D. et al. A Southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol. Biol. 1030, 151–161 (2013).
Parlanti, E., Fortini, P., Macpherson, P., Laval, J. & Dogliotti, E. Base excision repair of adenine/8-oxoguanine mispairs by an aphidicolin-sensitive DNA polymerase in human cell extracts. Oncogene 21, 5204–5212 (2002).
Thomas, D. C., Roberts, J. D. & Kunkel, T. A. Heteroduplex repair in extracts of human HeLa cells. J. Biol. Chem. 266, 3744–3751 (1991).
Zhao, X., Muller, E. G. & Rothstein, R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340 (1998).
Sells, M. A., Chen, M. L. & Acs, G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl Acad. Sci. USA 84, 1005–1009 (1987).
Kay, M. A., He, C. Y. & Chen, Z. Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 28, 1287–1289 (2010).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006 (2006).
Folco, E. G., Lei, H., Hsu, J. L. & Reed, R. Small-scale nuclear extracts for functional assays of gene-expression machineries. J. Vis. Exp. 64, e4140 (2012).
Ni, Y., Urban, S. & Hepatitis, B. Virus infection of HepaRG cells, HepaRG-hNTCP cells, and primary human hepatocytes. Methods Mol. Biol. 1540, 15–25 (2017).
Kaplan, P. M., Greenman, R. L., Gerin, J. L., Purcell, R. H. & Robinson, W. S. DNA polymerase associated with human hepatitis B antigen. J. Virol. 12, 995–1005 (1973).
Collaboration, O. R. The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat. Methods 13, 191–192 (2016).
Kadyrov, F. A. et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl Acad. Sci. USA 106, 8495–8500 (2009).
Acknowledgements
We thank X. Zhao (MSKCC) for sharing wild-type yeast stains and yeast-tagging plasmids; P. Modrich (Duke University) for sharing plasmids for expression of the RFC and POLδ complexes; B. Wan (MSKCC) for discussions on yeast-tagging strategies; the Zakian and Shenk lab (Princeton University) for sharing equipment and reagents for the yeast work; S. Port and S. Travis (Hughson lab, Princeton University), X. Gong (Yan lab, Princeton University), M. Estrella (Korennykh lab, Princeton University), R. Alfaro-Aco (Petry lab, Princeton University) and the Remus lab (MSKCC) for the their advice on protein purification and extract preparation; and B. Bratton and J. Sheehan (Gitai lab, Princeton University) for their advice on P1 phage transduction. We thank J. Gaska and other members of the Ploss laboratory for providing critical feedback on this manuscript. This work was supported in part by grants from the National Institutes of Health (grant no. R01 AI138797 to A.P.), a Research Scholar Award from the American Cancer Society (grant no. RSG-15-048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to A.P.) and funding from Princeton University. L.W. is a recipient of a postdoctoral fellowship award from the New Jersey Commission on Cancer Research (NJCCR, award no. AWD1005321, sponsor award no. DFHS17PPC011).
Author information
Authors and Affiliations
Contributions
The project was conceived by L.W. and A.P. All experiments were performed by L.W. All data were analysed by L.W. and A.P. The manuscript was written by L.W. and A.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characteristics of HBV rcDNA and effects of transfection of various rcDNA substrates to human hepatoma cells.
a, Four types of structures on HBV rcDNA need to be repaired by host repair factors to form cccDNA. Red line, a RNA primer/flap; Pol, HBV polymerase. A model describing the steps of rcDNA repair is as following: Removal of HBV Polymerase adduct from rcDNA can be achieved by TDP2, unknown endonuclease and protease digestion, generating 3 types (type A−type C) of deproteinated rcDNA (dp-rcDNA). Dp-rc DNA needs to be further processed to remove DNA flap and RNA primer/flap, fill the gap, and ligate the nicks. HBV rcDNA, and type A−type C dp-rcDNA can be mimicked by indicated recombinant rcDNA substrates (see Fig. 1 for details) to study cccDNA formation in our biochemical repair system. b−d, Examination of HBeAg production after transfection with X-tremeGENETM reagent of various HBV rcDNA substrates (described in (a) and Fig. 1a) into hNTCP HepG2 cells. b, Schematics of transfection experiments. 5 ng of individual control and rcDNA substrates together with 15 ng of mCherry expressing plasmids were transfected to hNTCP HepG2 cells seeded in a 96-well plate. 72 hours post transfection, transfection efficiency of each rcDNA substrates was estimated by mCherry fluorescence (c, scale bars indicate 400 μm) and HBeAg production in the supernatant was detected by ELISA (n=3, independent experiments) (d). Note that addition of NeutrAvidin strongly inhibited transfection efficiency shown in (c, compare 1st and 2nd panel). Statistic comparisons between ‘no rcDNA’ and ‘virion rcDNA’ samples were made using student-t test (two-tailed), *p value is 8x10-5. Bar value indicates mean of three measurements, and error bars are s.d. All data shown are representatives of three independent experiments.
Extended Data Fig. 2 Yeast cell extract fully supports repair of rcDNA to form cccDNA.
a, Repair of HBV virion-derived deproteinated rcDNA to cccDNA in yeast cell extract. Repair products are analyzed by Southern blotting (see methods for details). Un-treated recombinant rcDNA (RrcDNA) (lane 1), recombinant cccDNA (RcccDNA) (lane 2), and virion-derived rcDNA (lane 3) were used as controls. Lane 4, repair product following incubation of rcDNA with yeast cell extract (CE). L, linearized rcDNA. (b−c) Both minus and plus strands in RrcDNA are faithfully repaired by yeast cytoplasmic extract to form cccDNA. b, DNA sequences of the minus strand of repaired cccDNA (Fig. 2a, lane 3) and recombinant cccDNA. The 10 nt flap region on the minus strand of rcDNA is indicated by a blue shaded box and dashed lines. c, DNA sequences of the plus strand of repaired cccDNA (Fig. 2a, lane 3) and recombinant cccDNA. The ssDNA gap and RNA primer-containing region on the plus strand of rcDNA is indicated by a blue shaded box and dashed lines. All data shown are representatives of two independent experiments.
Extended Data Fig. 3 Schematic of a screening system using yeast cell extract to identify host factors essential for HBV rc- to cccDNA conversion.
Culture of yeast strains with a given protein ‘X’ tagged with Flag epitope are grown and pelleted. Cell pellets are treated with zymolyase to remove the cell wall and subsequently lysed. The cell lysate is separated into either cytoplasmic or nuclear fractions. Flag-tagged protein X is either mock depleted or depleted with anti-Flag antibody-conjugated beads. Both cell extracts are then incubated with recombinant rcDNA (RrcDNA), and the DNA from the repair reactions is subsequently extracted and separated on an agarose gel and visualized by EtBr staining or Southern blotting. A hypothetical gel pattern is shown – repair of RrcDNA will lead to the appearance of a cccDNA band that is expected to migrate faster. L stands for linearized RrcDNA due to nicking and shearing.
Extended Data Fig. 4 Tagging of yeast replication and repair factors with a C-terminal Flag epitope and effects of immuno-depletion of Pol1 and Pol2 on cccDNA formation.
a, Successful fusion of a 3x Flag tag at the endogenous chromosomal loci of various repair genes was confirmed by immunoblots using anti-Flag M2 antibody. Arrowheads indicate 3xFlag fusion gene products. Ponceau stains indicate equal loading. Repeated four times independently. b, Depletion of Pol1 (top) does not affect cccDNA formation (bottom). c, Immuno-depletion of Pol2 to undetectable levels (top) does not affect cccDNA formation (bottom). * indicates degraded proteins; ID stands for immunodepletion. Repeated twice independently.
Extended Data Fig. 5 Repair of rcDNA to form cccDNA using human nuclear extract.
a, Total cell, cytoplasmic, and nuclear extract from human HepG2 hepatoma cells were analyzed by western blotting to confirm separation of cytoplasmic and nuclear contents. Nuclear factor Lamin A/C, and cytoplasmic factor GAPDH were detected by specific antibodies. Cyto, cytoplasmic fraction; Nu, nuclear fraction. b, HBV virion-derived deproteinated rcDNA is repaired with human nuclear extract (NE) and cccDNA formation is analyzed by Southern blotting. Un-treated recombinant rcDNA (RrcDNA) (lane 1), recombinant cccDNA (RcccDNA) (lane 2), and virion-derived rcDNA (lane 4) were used as controls. Lane 3, repair product following incubation of rcDNA with human nuclear extract (NE). c, DNA sequences of the minus strand of repaired cccDNA (extracted from Fig. 3a, lane 8) and recombinant cccDNA. The 10 nt flap region on the minus strand of rcDNA is indicated by a blue shaded box and dashed lines. d, DNA sequences of the plus strand of repaired cccDNA (extracted from Fig. 3a, lane 8) and recombinant cccDNA. The ssDNA gap and RNA primer-containing region on the plus strand of rcDNA is indicated by a blue shaded box and dashed lines. All data shown are representatives of two independent experiments.
Extended Data Fig. 6 Replenishment of fresh nuclear extract at cccDNA formation plateau only marginally improves cccDNA formation.
Related to Fig. 3b, c. a, Schematics showing a time course for cccDNA formation reaction in human nuclear extracts. At 60 min, the reaction solution was split into two parts; one part was supplemented with a fresh aliquot of nuclear extract, while the other was not. b, Time course assay showing the kinetics of cccDNA formation from both RrcDNA (lanes 1−7) and NeutrAvidin (NA)-RrcDNA complex (lanes 8−14) as described in (a). Efficiency of cccDNA formation was calculated as in Fig. 2c and indicated in row ‘% repaired’. c, The efficiency of cccDNA formation from (b) is plotted against incubation time. All data shown are representatives of two independent experiments.
Extended Data Fig. 7 80% depletion of PCNA and POLD1 and 90% depletion of FEN-1 do not affect cccDNA formation in human cytoplasmic extracts.
a, Depletion of 80% of PCNA in human cytoplasmic extract (left) does not affect repair of RrcDNA (middle) and NA-RrcDNA21. Experiments were carried out as in Fig. 3g–i. M, Mock depletion using mouse IgG. b−c, same as (a), except that POLD1 and FEN-1 are examined, respectively. M, Mock depletion using rabbit IgG. d, Antibodies against the RFC4 subunit of the RFC complex and LIG1 fail to achieve protein depletion in human cytoplasmic extracts. (a−d) a relative volume (rel. vol) of 100 corresponds to 0.5 μl extract. All data shown are representatives of two independent experiments.
Extended Data Fig. 8 Estimation of the concentration of repair factors in human cell extract.
a, The concentrations of PCNA in cytoplasmic extract (CE, top) and nuclear extract (NE, bottom) are compared by immunoblot with purified 6xhistidine-tagged PCNA of the indicated amount. 1 μl of extracts were contained in CE and NE. Relative signal of PCNA bands was calculated by setting that of cytoplasmic extract to 1. b−e, Same as (a), except that RFC4, LIG1, POLD1, and FEN-1 are examined. LIG1-p, phosphorylated form of LIG1. λPP, lambda phosphatase. Note that λPP treatment (lane 2) in (c) reduces the mobility of phosphorylated LIG1 (lane 1) to that of the non-phosphorylated form (lanes 2−4). All data shown are representatives of two independent experiments.
Extended Data Fig. 9 All five factors are necessary and sufficient for repair of virion derived rcDNA and a summary of human factors involved in cccDNA formation in this study.
a, The repair of virion derived type C rcDNA requires all five factors as recombinant rcDNA substrates used in Fig. 4e. b, The core components involved in DNA lagging strand synthesis – PCNA, RFC, POLδ, FEN-1 and LIG1 – constitute a minimal set of factors for cccDNA formation in this study. All data shown are representatives of two independent experiments.
Extended Data Fig. 10 Strains and plasmids used in this study.
Saccharomyces cerevisiae strains were isogenic to W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535)27. Only one strain is listed for each genotype, but at least two independent isolates of each genotype were used in the experiments.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed agarose gels and Southern blots.
Source Data Fig. 2
Unprocessed agarose gels and western blots.
Source Data Fig. 2
Numerical source data.
Source Data Fig. 3
Unprocessed agarose gels and western blots.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Unprocessed protein gels, agarose gels and Southern blots.
Source Data Fig. 4
Numerical source data.
Source Data Fig. 5
Unprocessed agarose gels.
Source Data Fig. 6
Unprocessed agarose gels and Southern blots.
Source Data Fig. 6
Numerical source data.
Source Data Extended Data Fig. 1
Numerical source data.
Source Data Extended Data Fig. 2
Unprocessed Southern blots.
Source Data Extended Data Fig. 4
Unprocessed agarose gels and western blots.
Source Data Extended Data Fig. 4
Numerical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots and Southern blots.
Source Data Extended Data Fig. 6
Unprocessed agarose gel.
Source Data Extended Data Fig. 6
Numerical source data.
Source Data Extended Data Fig. 7
Unprocessed agarose gels and western blots.
Source Data Extended Data Fig. 7
Numerical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed Southern blots.
Rights and permissions
About this article
Cite this article
Wei, L., Ploss, A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation. Nat Microbiol 5, 715–726 (2020). https://doi.org/10.1038/s41564-020-0678-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0678-0
This article is cited by
-
The scientific basis of combination therapy for chronic hepatitis B functional cure
Nature Reviews Gastroenterology & Hepatology (2023)
-
Long-term hepatitis B virus infection of rhesus macaques requires suppression of host immunity
Nature Communications (2022)
-
PRKDC promotes hepatitis B virus transcription through enhancing the binding of RNA Pol II to cccDNA
Cell Death & Disease (2022)
-
Circadian control of hepatitis B virus replication
Nature Communications (2021)
-
Hepatitis B virus cccDNA is formed through distinct repair processes of each strand
Nature Communications (2021)