ABSTRACT
Enhancer II (ENII) is one of the critical cis-elements in the Hepatitis B Virus (HBV) genome for the hepatic viral gene transcription and DNA replication. The liver-specific activity of ENII is regulated by multiple liver-enriched transcription factors, including LRH-1/hB1F, HNF1, HNF3β, HNF4 and C/EBP. Knowledge on the interplay of these important factors is still limited. In this study, we demonstrate a functional synergism between the orphan nuclear receptor LRH-1/hB1F and the homeoprotein HNF1 in up-regulating the liver-specific activity of ENII. This synergism is sufficient for initiating the viral gene transcription and DNA replication in non-hepatic cells. We have defined the activation domains in hB1F and HNF1 that contribute to the synergism. We further show that hB1F and HNF1 can interact directly in vitro and have mapped the domains required for this interaction.
Similar content being viewed by others
INTRODUCTION
Infection with hepatitis B virus (HBV) is highly restricted to hepatocytes. The remarkable liver tropism is likely due to both the hepatic expression of a yet-to-be identified viral receptor(s) and the control of viral mRNA synthesis and DNA replication by liver-enriched regulators. Enhancer II (ENII) is one of the critical cis-elements in the HBV genome for the hepatic viral gene transcription and DNA replication by modulating the core promoter (Cp) activity1, 2. The liver-specific activity.of ENII is regulated by multiple liver-enriched transcription factors, including hB1F, HNF1, HNF3β, HNF4, and C/EBP3, 4, 5, 6, 7, 8, 9. The interaction bewteen each of these factors and ENII has been studied in great detail, but less is known on the interplay of these important regulators, which we think is crucial for the function of ENII as well as the life cycle of HBV.
In previous studies, we showed that the orphan nuclear receptor hB1F and homeoprotein HNF1 can bind to the B1 and B2 elements of ENII respectively and stimulate its liver-specific activity, which consequently regulates hepatic viral gene transcription and DNA replication 3, 9, 10. hB1F (also known as LRH-1, FTF, CPF) is a human homolog of the Drosophila fushi tarazu factor I (FTZ-F1)3, 11. It shares a common modular structure with other members of the nuclear receptor superfamily, including the N-terminal A/B domain, a zinc finger DNA binding domain (DBD), a hinge region and a ligand binding domain (LBD). With a POU box located at the amino-terminus, hepatocyte nuclear factor 1(HNF1) belongs to the POU homeoprotein subfamily12, 13. hB1F and HNF1 are key regulators of many liver-specific genes. Our preliminary results suggested that hB1F and HNF1 act synergistically in regulating the activity of ENII since the co-expression of both factorS cause a dramatic activation of ENII that was much stronger than the additive effects result from the expression of either factor alone14. In the present study, the functional synergism between hB1F and HNF1 in up-regulating the activity of ENII was explored in detail. We demonstrate that the synergistical interplay between hB1F and HNF1 is crucial for hepatic HBV gene transcription and DNA replication. We then map the activation domains in hB1F and HNF1 for the functional synergism and further show that hB1F and HNF1 are able to interact directly.
MATERIALS AND METHODS
Plasmid construction
Plasmids in CAT assays
Primers for the plasmid construction are listed in Tab 1. The reporter pABCDCAT containing the HBV ENII/Cp region located upstream from the CAT reporter gene has been described3.
The pCMVhB1F contains the full length hB1F cDNA cloned in the pCMVpoly vector3, 14. The pCMVhB1FDAF2 that lacks the activation function 2 (AF2) of hB1F was made by treating the pCMVhB1F sequentially with BamHI/StyI, Klenow enzyme and T4 DNA ligase. To generate the pCMVhB1FDRII, the upstream and downstream fragments flanking the RII region (aa301-345) in the LBD of hB1F were amplified with primer pairs P1/P2 and P3/P4, followed by the digestion with ApaI/EcoRI and EcoRI/NsiI respectively, and used to replace the ApaI/NsiI fragment in the pCMVhB1F. The pCMVhB1FDRIII and pCMVhB1FDR(II+III) were similarly constructed, using different combinations of primer pairs to amplify upstream and downstream fragments flanking the RIII (aa392-414) or R(II+III) (aa301-414) region in the LBD of hB1F. Primer pairs P1/P5 and P6/P4 were used for the pCMVhB1FDRIII while P1/P2 and P6/P4 for the pCMVhB1FDR(II+III). The pCMVhB1FN1 was made by subcloning the EcoRI fragment from the pBShB1FN1 (see below) in the pCMVpoly vector.
The pRSVHNF1 or pRSVHNF1DBD contain the full-length or DNA binding domain (aa1-281) of HNF1 respectively3, 14. The pRSVHNF1ADI was made by treating pRSVHNF1 with BalI/StuI and T4 DNA ligase. To construct the pRSVHNF1DADI, the pACTHNF114 was treated sequentially with BamHI, Klenow enzyme and SstI to release an 1.6 kb fragment that was used to replace the SstI/StuI fragment in the pRSVHNF1.
Plasmids for in vitro translation
The pcDNA3HNF1 and pBShB1F contain the full-length HNF1 and hB1F cDNAs cloned in the pcDNA3 (Invitrogen) and pBS(+) (Stratagene) respectively 14. The pBShB1FN1, pBShB-1FN2, pBShB1FC encode aa1-177, aa1-138, aa157-495 of hB1F in the pBS (+) respectively. The pBShB1FN1 was made by treating the pBShB1F sequentially with StuI/StyI, klenow enzyme and T4 DNA ligase. To generate the pBShB1FN2, hB1F fragment was amplified with primers P7 and P8, followed by digestion with BglII/BamHI and used to replace the BglII/BamHI fragment in the pBShB1F. To generate the pBShB1FC, hB1F fragment was amplified with primers P9 and P10, followed by digestion with EcoRI/PstI and inserted into the pBS(+).
Plasmids of GST fusion proteins
The pGSTHNF1FL contains the full-length HNF1 cDNA fused to the GST protein has been described3, 14. The pGSThB1FDBD contains the DBD of hB1F (aa35-215) fused to the GST protein in the pGEX-3x (Amersham)3, 14. The pGSTHNF1N, pGSTHNF1C, pGSTHNF1M, pGSTHNF1DBD and pGSTHNF1- homeo contain PCR-amplified fragments corresponding to aa1-263, aa264-628, aa128-302, aa1-281 and aa198-281 of HNF1 respectively fused to the GST protein in the pGEX-3x.
Transient transfection and CAT assay
Human hepatoma HepG2 and cervical carcinoma HeLa cells were maintained in Dulbecco?s modified Eagle?s medium (DMEM) supplemented with 10% fetal bovine serum. HeLa cells on 60-mm dishes were transfected by the calcium phosphate coprecipitation method typically with 1 μg of the pABCDCAT reporter, 1 μg of an expression plasmid or an empty vector and 2 μg of the pCMVLacZ (an internal control encoding β-galactosidase, Promega). Cells were harvested at 48 h post-transfection and lysed with three rounds of freeze-thraw in 100 μl of 0.25 M Tris-HCl (pH 7.5). The β-galactosidase activity in the cell lysates was measured according to a standard protocol15 and served as an internal control for normalizing the transfection efficiency. The amount of the cell lysates that would be used in the CAT assays were adjusted proportionally based on the β-galactosidase activities. CAT assays of cell lysates were performed according to a published protocol16. Each experiment was performed in duplicate dishes and repeated at least three times.
Analysis of viral RNA and DNA
HeLa cells grown on 60-mm dishes were transfected with 4 μg of the pWT17 that contains a terminally redundant, replication competent HBV genome (adr), 4 μg of an expression plasmid or the empty vector and 1 μg of the pCMVLacZ. Three days post-transfection, transfection, cells were harvested for viral RNA and DNA analysis. Total RNA was isolated with TRIzol reagent (Gibco) according to the manufacturer?s instruction. For Northern blot analysis, 20 μg of total RNA was electrophoresed on 1% denaturing agarose gel and transferred onto Hybond N+ membrane (Amersham). For southern analysis, viral replicative DNA intermediates associated with intracellular core particles were prepared as described previously18. DNA was subject to electrophoresis on agarose gels and transferred onto Hybond N+ membrane. The Membranes were hybridized with an 1.5 kb XbaI fragment of HBV DNA19 labeled with the prime-agene labeling system (promega) according to the manufacturer?s protocol. Signals were detected by autoradiography. RNA or DNA from HepG2 cells transfected with the pWT was run alongside the samples from HeLa cells and served as positive controls 17.
Expression of GST fusion proteins
3 ml 1:100 dilution of overnight cultures of E.coli BL21(DE3) cells containing GST fusion constructs were incubated for 2 hours at 37 °C and then induced with IPTG for 1.5 h at 30 °C (pGSTHNF1FL) or 37 °C. To achieve optimal expression of fusion proteins, different IPTG concentrations were used for different constructs (pGSTHNF1FL, 0.02 mM; pGSTHNF1C, 0.05 mM; pGSTHNF1DBD, pGSTHNF1N, pGSTHNF1M, pGSTHNF1-homeo and pGS-ThB1-FDBD, 0.1 mM). GST fusion proteins were purified as described14. Protein concentrations were determined by comparing sample lanes to a BSA standard on Coomassie blue stained SDS-gels.
In vitro translation
Full-length or fragments of hB1F and HNF1 were in vitro translated using TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instruction with [35S] methionine (Amersham) included in the reaction mix to label the translated proteins. 2 μl of translated proteins were subject to SDS-PAGE, followed by autoradiography. Relative concentrations of various in vitro translated products were determined based on the radioactive signal intensities.
GST pull-down assay
Equal amount of in vitro translated proteins were mixed respectively with 5 μg of a GST fusion protein conjugated on 50 μl of glutathione beads, pre-incubated at 4 °C in 150 μl of binding buffer (100 mM KCl, 20 mM Hepes pH 7.9, 0.1% NP-40, 5 mM MgCl2, 0.2% BSA, 10% glycerol, 0.1 mM PMSF). The binding reaction was allowed to proceed for 1.5 h at 4°C. After extensive washing with the wash buffer (binding buffer without BSA and glycerol), the beads were resuspended in 50 μl of SDS loading buffer and subject to SDS-PAGE. The proteins were visualized by autoradiography. in vitro translated [35S] methionine-labeled luciferase served as a non-specific control.
RESULTS
hB1F and HNF1 synergistically up-regulate the activity of HBV ENII
HeLa cells that lack the endogenous expression of hB1F 3 and HNF1 20 were used to study the stimulationon ENII by hB1F and HNF1. Cells were co-transfected with the expression plasmid of hB1F and/or HNF1 and the reporter pABCDCAT. The pCMVLacZ was included and served as an internal control (see Materials and Methods). As expected, the reporter pABCD-CAT containing the ENII and Cp of HBV (Fig 1A)21 was inactive in non-hepatic HeLa cells. The exogenous expression of either hB1F or HNF1 only resulted in a weak stimulation (∼3-fold). However, the co-expression of the exogenous hB1F and HNF1 activated the reporter significantly up to 25-fold (Fig 1B). This result clearly demonstrated that the up-regulation of the ENII activity was a synergistical action by hB1F and HNF1.
hB1F and HNF1 synergistically up-regulate the activity of HBV ENII. (A) The ENII and Cp of HBV. ENII contains functional elements, A, B1, B2 and B3. The binding sites of hB1F and HNF1 were underlined. Cp overlaps with ENII, spanning from B3 to D. (B) HeLa cells was co-transfected with 1 μg of pABCDCAT, 1 μg of an indicated expression plasmid or the empty vector and 2 μg of pCMVLacZ. Transfection efficiencies were normalized according to the β-galactosidase activities (see materials and methods and reference 15). The CAT activity of pABCDCAT was taken as 1. The stimulations on the reporter by hB1F and/or HNF1 were shown as fold of induction. Data from at least three independent experiments were presented. A representative photo of the CAT assay was shown.
hB1F/HNF1 synergism is crucial for viral gene transcription and DNA replication
Does the synergism between hB1F and HNF1 indeed play an important role in HBV gene transcription and DNA replication? To answer this question, transient cotransfection assays were performed in HeLa cells with the pWT and the expression plasmid of hB1F and/or HNF1. The pWT contains a terminally redundent, replication- competent HBV genome capable of generating all the viral mRNAs and proteins under the control of viral promoters and enhancers in hepatocytes but not in HeLa cells (Fig 2A)17. We chose HeLa cells over hepatic cells to avoid the interference by endogenous liver-enriched factors present in the latter. A similar approach was taken to examine the role of nuclear receptors in the regulation of HBV replication22.
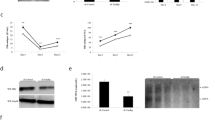
hB1F/HNF1 synergism on viral gene transcription and DNA replication. (A) Schematic representation of the terminally redundant, replication-competent HBV genome in the pWT. HBV transcripts were depicted by arrowed lines. (B) Northern blot of viral RNAs. 20 mg of total RNAs from HeLa cells transfected with pWT or cotransfected with pWT and expression plasmids of hB1F and/or HNF1 were hybridized with an HBV DNA probe. The 3.5 kb represents the precore/pregenomic viral RNA and the 2.4/2.1 kb the preS/S mRNA. RNAs from HepG2 cells transfected with pWT was run alongside as a positive control. (C) Southern blot of viral replication intermediates. HBV DNA in the cellular viral nucleocapsids was isolated from transfected HeLa cells and hybridized with the same HBV DNA probe used in the northern blot. Viral DNA from HepG2 cells transfected with pWT was run alongside as a positive control. ss, single strand HBV DNA; oc, open circular HBV DNA. Transfection efficiencies were normalized by co-transfection with an internal control plasmid pCMVLac that encodes β-galactosidase as described in the material and methods.
Total RNA was prepared 3 days post-transfection and subject to northern blot to analyze the synthesis of viral RNAs. Two major viral transcripts, the 3.5kb precore/ pregenomic RNA and the 2.4/2.1kb preS/S mRNA 17, 18, 19, 22 were clearly detected in HepG2 cells (Fig 2B, lane 5). In contrast, Viral RNAs were undetectable in HeLa cells transfected with the pWT alone (Fig 2B, lane 1). Expression of hB1F did not stimulate viral gene transcription (Fig 2B, lane 2) while expression of HNF1 resulted in a weak but apparent increase in both the 2.1/2. 4kb and 3.5kb viral transcripts (Fig 2B, lane 3). This is probably because HNF1 can stimulate multiple viral ciselements besides ENII, including the large surface antigen promoter and enhancer I (ENI)23. In contrast, viral gene transcription was significantly enhanced by the co-expression of hB1F and HNF1 in HeLa cells (Fig 2B lane 4). Therefore, the synergism between hB1F and HNF1 is functionally crucial for viral gene transcription.
To analyze the up-regulation of viral DNA replication by hB1F and HNF1, replication intermediates associated with the intracellular core particles were prepared from transfected HeLa cells and subject to southern blot. Replication intermediates were undetectable in cells transfected with the pWT or co-transfected with either hB1F or HNF1 expression plasmid (Fig 2C, lane 1-3), indicating that neither hB1F nor HNF1 alone is sufficient for initiating viral DNA replication in HeLa cells under the experimental condition used in this study. In contrast, viral replication intermediates could be readily detected in HeLa cells co-transfected with both hB1F and HNF1 expression plasmids (Fig 2C, lane 4). This result clearly demonstrated that the synergism between hB1F and HNF1 is crucial and sufficient for initiating viral DNA replication in non-hepatic cells. However, it is noteworthy that the replication intermediate in these cells is exclusively the single-stranded form. The reason for the absence of the partially double-stranded form (ocDNA) (Fig 2C, lane 5) which represents the mature viral genome in infectious virions are currently unknown.
Activation domains involved in hB1F/HNF1 synergism
Both hB1F and HNF1 have a modular structure, including DNA binding domains for interacting with target DNA and transactivation domains for regulating gene transcription. To find out the domains involved in the synergism between hB1F and HNF1, a series of hB1F and HNF1 deletion constructs (Fig 3A and B) were generated and analyzed in co-transfection assays for their capabilities of synergistically stimulating the activity of ENII. As shown in Fig 3A, the DBD of HNF1 (HNF1- DBD) did not show any synergism with the full-length hB1F. Likewise, the DBD of hB1F (hB1FN1) also failed to display any synergism with the full-length HNF1 (Fig 3B), indicating that the DBD alone is not sufficient for the synergism between hB1F and HNF1.
Determination of activation domains required for hB1F HNF1 synergism. (A) (left panel) Schematic presentation of HNF1 deletion constructs. (right panel) Effects of HNF1 deletions on hB1F/HNF1 synergism. The transcriptional activations of ENII by various HNF1 deletion mutants were investigated through co-transfection assays performed in HeLa cells with 1 μg of pABCDCAT, 1 μg of indicated HNF1 expression plasmids (black bars). To examine the synergism between hB1F and each HNF1 deletion mutant, 1 μg of hB1F expression plasmid was included in the co-transfection in addition to pABCDCAT and each HNF1 construct (gray bars). (B) (left panel) Schematic presentation of hB1F deletion constructs. (right panel) Effects of hB1F deletions on hB1F/HNF1 synergism. The transcriptional activations of ENII by various hB1F deletion mutants were investigated through co-transfection assays performed in HeLa cells with 1 μg of pABCDCAT, 1 μg of indicated hB1F expression plasmids (black bars). To examine the synergism between HNF1 and each hB1F deletion mutant, 1 μg of HNF1 expression plasmid was included in the co-transfection in addition to pABCDCAT and each hB1F construct (gray bars). Transfection efficiencies were normalized by co-transfection with pCMVLac that encodes β-galactosidase as described in the material and methods. The CAT activity of pABCDCAT was taken as 1. Data from at least three independent experiments were presented.
Since the activation domains of hB1F and HNF1 are composed of multiple subdomains, it is possible that individual subdomain may not contribute equally to the synergism between hB1F and HNF1. Therefore, subdomain deletion constructs were generated to investigate which individual activation subdomain was required for the synergism. HNF1ADI with an intact activation subdomain I (ADI)24, 25 but without the activation subdomains II (ADII) and III (ADIII), retained the activator function but lost the synergism with hB1F, only displaying an additive stimulation (Fig 3A). In contrast, HNF1δADI that lacked the ADI was almost inert in transcriptional activation, nevertheless it still retained an obvious synergism with hB1F (Fig 3A). The above data suggested that the N-terminal of the HNF1 activation domain (aa282-546) including the ADII and ADIII (but not ADI) is involved in the synergism between hB1F and HNF1. Reciprocally, three conserved subdomains activation in the LBD of hB1F, namely region II (RII), region III (RIII) and AF-23, were deleted either alone or in combination to define the regions required for the synergism. As shown in Fig 3B, all the mutants lost the transcriptioncapability as well as the synergism with HNF1, indi cating that the RII, RIII, and AF-2 are indispensable for the synergism.
hB1F and HNF1 interact directly
It is possible that hB1F and HNF1 can interact directly. To test this possibility, we performed GST pull-down experiments with GST-HNF1 fusion proteins (Fig 4A) and in vitro translated [35S] methionine-labeled hB1F. As shown in Fig 4B, the full-length hB1F could directly interact with the full-length as well as the DBD of HNF1 (HNF1FL and HNF1DBD, lane 3 and 6). Only weak or no interaction was observed between hB1F and the Nor C-terminal truncated HNF1 (HNF1N and HNF1C, lane 4 and 5), suggesting that the boundary region joining HNF1N and HNF1C is required for the interaction betweenHNF1N and HNF1C is required for the interaction between hB1F and HNF1. This speculation was confirmed by the strong interaction between hB1F and HNF1M that spans the boundary region (lane 8).indicating that the DBD of hB1F alone was sufficient for Additional construct showed that the homeodomain of HNF1 (aa198-281) was sufficient to mediate the interaction with hB1F (HNF1homeo, lane 7). Reciprocally, domains of hB1F required for the interaction with HNF1 were similarly identified by GST pull-down assays. As shown in Fig 4D, the full-length hB1F, hB1FN1 and hB1FN2 but not hB1FC could interact with the full-length HNF1, indicating that the N-terminal region of hB1F including the A/B domain and DBD were involved in the interaction. To elucidate the precise domain of hB1F for the interaction, GST-hB1FDBD, which only contains the DBD of hB1F was incubated with labeled in vitro translated full-length HNF1. As shown in Fig 4E, hB1- FDBDcould interact with the full-length HNF1, the interaction with HNF1. Taken together, the above results demonstrated that the homeodomain of HNF1 and the DBD of hB1F were involved in the direct interaction between these two factors in vitro.
Interaction of hB1F with HNF1 in vitro. (A) Schematic presentation of GST-HNF1 fusion proteins. (B) The homeodomain of HNF1 interacts with hB1F. In vitro translated and [35S] labeled full length hB1F was incubated with 5 μg of GST or various GST-HNF1 fusion proteins immobilized on the glutathione beads. hB1F proteins retained on the beads after extensive wash were analyzed by SDS-PAGE and autora diography. Input, 25% of the in vitro translated hB1F used in the GST pull-down assay. (C) Schematic presentation of GST-hB1F fusion proteins. (D) N terminal part of hB1F mediates hB1F/HNF1 interaction. In vitro translated and [35S] labeled full length or truncated hB1Fs were incubated with 5 μg of GST or GST-HNF1FL immobilized on glutathione beads. hB1F proteins retained on the beads after extensive wash were analyzed by SDS-PAGE and autoradiography. Input, 25% of the in vitro translated hB1F used in the GST pull-down assay. (E) DBD of hB1F is sufficient for the interaction between hB1F and HNF1. Labeled HNF1 or luciferase was synthesized by in vitro translation and incubated with GST or GST-hB1FDBD immobilized on glutathione beads. Input contains 25% of in vitro translated proteins used in the GST pull-down assay.
DISCUSSION
hB1F and HNF1 are two of the multiple liver enriched transcriptional factors that have been implicated in the regulation of the liver specific activity of HBV ENII. The importance of these two factors to the function of ENII is well established3, 4, 5, 10. However, either factor alone is not able to fully initiate viral gene transcription and replication in non-hepatic cells. In this study, we showed that hB1F and HNF1 act synergistically in the up-regulation of the liver specific activity of ENII. We further demonstrated that the synergistical interplay between hB1F and HNF1 was sufficient for initiating viral gene transcription and replication in non-hepatic cells. Our data provide the first experimental evidence that the synergism between hB1F and HNF1 might define a major determinant of the liver tropism of ENII.
The mapping of the required activation domains in hB1F and HNF1 for the synergism between these two factors revealed some intriguing aspects of the synergism. One mutant HNF1 that contains an internal deletion of the ADII and ADIII is capable of stimulating ENII but fails to synergize with hB1F. In contrast, another mutant HNF1 that contains the ADII and ADIII but lacks the ADI is inert in transactivation, yet able to synergize with hB1F. This result not only indicates that the ADII and ADIII of HNF1 are required for the synergism but also implicates that at least on the ENII/Cp reporter, the synergism between hB1F and HNF1 does not necessarily require a transcriptional active HNF1. With respect to hB1F, since all hB1F mutants with the deletions in the LBD are inert in transactivation as well as synergism with HNF1, it is impossible from this study to deduce whether the synergism requires a transcriptional active hB1F.
The in vitro binding assay revealed that the DBD of hB1F and the homeodomain of HNF1 might mediate the direct interaction between these two factors. It awaits further experiment to determine whether hB1F and HNF1 interact in vivo and these two domains are sufficient for the interaction between these two factors. Nevertheless, from the data in this study, it is clear that the synergism of hB1F and HNF1 require more than a physical interaction between the two factors. As above-mentioned, at least the ADII and ADIII of HNF1 and the LBD of hB1F are also involved in the synergism. It has been well documented that the LBD in nuclear receptors can interact various cofactors26, 27 such as the steroid receptor coactivator-1 (SRC-1) and the related glucocorticoid receptor activation protein-1 (GRIP-1). On the other hand, many cofactors, including CBP, P/CAF and SRC-1 can interact with distinct activation regions of HNF128. The requirement of the ADII and ADIII of HNF1 and the LBD of hB1F suggests that some cofactors might be crucial for the synergism between hB1F and HNF1. Future investigation should elucidate these important cofactors.
It is noteworthy that both the homeodomain of HNF1 and the DBD of hB1F are highly conserved structural modules found in almost all the members in the homeoprotein and nuclear receptor superfamilies. It is thus tempting to speculate that the interaction between homeoproteins and nuclear receptors may occur frequently, which probably has important biological functions in vivo. Kakizawa et al29 describes a functional interaction between the homeodomain of Oct-1 and the DBD of retinoid X receptor. Our findings presented another example of a functional interaction between a homeoprotein and a nuclear receptor. Together they suggest that the functional interplay between homeoproteins and nuclear receptors may extend to other members of these two important families of transcriptional factors, which should be an interesting issue for future investigation.
Abbreviations
- hB1F:
-
hepatitis B virus enhancer II B1 binding factor
- LRH-1:
-
liver receptor homologue-1
- HNF1:
-
hepatocyte nuclear factor 1
- HBV:
-
hepatitis B virus
- Cp:
-
core promoter
- ENII:
-
enhancer II
- AD:
-
activation domain
- DBD:
-
DNA binding domain
- LBD:
-
ligand binding domain
References
Wang Y, Chen P, Wu X, et al. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol 1990; 64:3977–81.
Yee JK . A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science 1989; 246:658–61.
Li M, Xie YH, Kong YY, Wu X, Zhu L, Wang Y . Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem 1998; 273:29022–31.
Gilbert S, Galarneau L, Lamontagne A, Roy S, Belanger L . The hepatitis B virus core promoter is strongly activated by the liver nuclear receptor fetoprotein transcription factor or by ectopically expressed steroidogenic factor 1. J Virol 2000; 74:5032–9.
Li M, Xie YH, Wu X, Wang Y . HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology 1995; 214:371–8.
Guo W, Chen M, Yen TS, Ou JH . Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol 1993; 13:443–8.
Lopez-Cabrera M, Letovsky J, Hu KQ, Siddiqui A . Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA 1990; 87:5069–73.
Raney AK, Johnson JL, Palmer CN, McLachlan A . Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol 1997; 71:1058–71.
Wang WX, Li M, Wu X, Wang Y, Li ZP . HNF1 is critical for the liver-specific function of HBV enhancer II. Res Virol 1998; 149:99–108.
Xie Y, Li M, Wang Y, Hofschneider PH, Weiss L . Site specific mutation of the hepatitis B virus enhancer II B1 element: effect on virus transcription and replication. J Gen Virol 2001; 82: 531–5.
Nitta M, Ku S, Brown C, Okamoto AY, Shan B . CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci USA 1999; 96:6660–5.
Frain M, Swart G, Monaci P, Nicosia A, Staempfli S, Frank R, Cortese R . The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell 1989; 59:145–57.
Baumhueter S, Mendel DB, Conley PB, et al. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev 1990; 4:72–9.
Cai YN, Li M, Zhou Q, Kong YY, Wang Y . hB1F and HNF1 work synergistically in the regulation of the activity of HBV ENII. Chinese Science Bulletin 2000; 45:35–9.
Sambrook J, Fritsch EF, Maniatis T . Molecular cloning, cold spring harbor laboratory, cold spring harbor, New York, 1989.
Gorman CM, Moffat LF, Howard BH . Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol 1982; 2:1044–51.
Yu XM, Mertz JE . Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J Virol 2001; 75:11354–64.
Fu L, Cheng YC . Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L (-) SddC (3TC) resistance. Biochem. Pharmacol 1998; 55:1567–72.
Feng Y, Kong YY, Wang Y, Qi GR . Inhibition of hepatitis B virus by hammerhead ribozyme targeted to the poly(A) signal sequence in cultured cells. Biol Chem. 2001; 382:655–60.
Cereghini S, Yaniv M, Cortese R . Hepatocyte dedifferentiation and extinction is accompanied by a block in the synthesis of mRNA coding for the transcription factor HNF1/LGB1. EMBO J 1990; 9:2257–63.
Wu X, Zhu L, Li ZP, Koshy R, Wang Y . Functional organization of Enhancer (ENII) of hepatitis B Virus. Virology 1992; 191:490–4.
Tang H, McLachlan A . Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci USA 2001; 98:1841–6.
Raney AK, Easton AJ, Milich DR, McLachlan A . Promoterspecific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. J Virol 1991; 65:5774–81.
Nicosia A, Monaci P, Tomei L, et al. A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell 1990; 61:1225–36.
Bach I, Yaniv M . More potent transcriptional activators or a transdominant inhibitor of the HNF1 homeoprotein family are generated by alternative RNA processing. EMBO J 1993; 12:4229–42.
Moras D, Gronemeyer H . The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol 1998; 10:384–91.
Torchia J, Glass C, Rosenfeld MG . Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol 1998; 10:373–83.
Soutoglou E, Papafotiou G, Katrakili N, Talianidis I . Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J Biol Chem. 2000; 275:12515–20.
Kakizawa T, Miyamoto T, Ichikawa K, et al. Functional interaction between Oct-1 and retinoid X receptor. J Biol Chem 1999; 274:19103–8.
Acknowledgements
The authors are grateful to Dr. E Lai for the pCMVpoly vector, Dr. M YANIV for the HNF1 expression plasmid and Dr. XM Yu for the pWT. This work is supported by the National Natural Science Foundation of China (30100088), High Technology Research and Development Project (2001-AA221261) and Basic Research Program from Ministry of Science and Technology (G19990 54105). Y Xie is supported by a Qi Ming Xing program (01QA14046) from Shanghai Science and Technology Committee. M Li is supported by a fellowship for outstanding Ph.D students from the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
CAI, Y., ZHOU, Q., KONG, Y. et al. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res 13, 451–458 (2003). https://doi.org/10.1038/sj.cr.7290187
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290187
Keywords
This article is cited by
-
DeepHBV: a deep learning model to predict hepatitis B virus (HBV) integration sites
BMC Ecology and Evolution (2021)
-
Genome-wide association study of prevalent and persistent cervical high-risk human papillomavirus (HPV) infection
BMC Medical Genetics (2020)
-
Integrated Analysis of Copy Number Variations and Gene Expression Profiling in Hepatocellular carcinoma
Scientific Reports (2017)