Postprandial Circulating miRNAs in Response to a Dietary Fat Challenge
Abstract
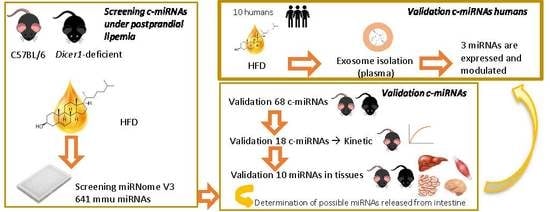
:1. Introduction
2. Materials and Methods
2.1. Animal Models
2.2. High-Fat Dietary Challenge
2.3. Sample Collection
2.4. Plasma miRNAs Determination
2.4.1. C-miRNA Screening
2.4.2. Individual C-miRNA Analysis
2.4.3. Analysis of miRNAs in Tissues
2.5. Human C-miRNA Validation
2.5.1. Human Study
2.5.2. Exosome Isolation
2.5.3. Biochemical Analysis
2.6. Bioinformatic Analysis
2.7. Statistical Study
3. Results and Discussion
3.1. miRNA Screening in Response to Dietary Fat Challenge
3.2. Validation of Modulated miRNAs
3.3. Kinetic of Validated miRNAs
3.4. Tissue Expression of miRNAs
3.5. Human miRNA Expression
3.6. Target Genes and Predictive Functionality of Postprandially Modulated miRNAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, S.-S.; Jin, J.-P.; Wang, J.-Q.; Zhang, Z.-G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Lawler, P.R.; Akinkuolie, A.O.; Harada, P.; Glynn, R.J.; Chasman, D.I.; Ridker, P.M.; Mora, S. Residual Risk of Atherosclerotic Cardiovascular Events in Relation to Reductions in Very-Low-Density Lipoproteins. J. Am. Hear. Assoc. 2017, 6, e007402. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, A.C.; Musunuru, K. Treatment of Dyslipidemia Using CRISPR/Cas9 Genome Editing. Curr. Atheroscler. Rep. 2017, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Burton-Freeman, B. Postprandial metabolic events and fruit-derived phenolics: A review of the science. Br. J. Nutr. 2010, 104, S1–S14. [Google Scholar] [CrossRef]
- Nakamura, K.; Miyoshi, T.; Yunoki, K.; Ito, H. Postprandial hyperlipidemia as a potential residual risk factor. J. Cardiol. 2016, 67, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Kats, D.; Sharrett, A.R.; Ginsberg, H.N.; Nambi, V.; Ballantyne, C.M.; Hoogeveen, R.C.; Heiss, G. Postprandial lipemia and the risk of coronary heart disease and stroke: The Atherosclerosis Risk in Communities (ARIC) Study. BMJ Open Diabetes Res. Care 2017, 5, e000335. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.B.; Moughan, P.J.; Wood, L.G.; Singh, H.; Garg, M.L. Postprandial lipemia: Factoring in lipemic response for ranking foods for their healthiness. Lipids Health Dis. 2017, 16, 178. [Google Scholar] [CrossRef]
- Hazim, J.; Hlais, S.; Ghattas, H.; Shatila, D.; Bassil, M.; Obeid, O. Phosphorus supplement alters postprandial lipemia of healthy male subjects: A pilot cross-over trial. Lipids Health Dis. 2014, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Novák, J.; Olejníčková, V.; Tkáčová, N.; Santulli, G. Mechanistic Role of MicroRNAs in Coupling Lipid Metabolism and Atherosclerosis. Adv. Exp. Med. Biol. 2015, 887, 79–100. [Google Scholar] [Green Version]
- Gambardella, J.; Santulli, G. Integrating diet and inflammation to calculate cardiovascular risk. Atherosclerosis 2016, 253, 258–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuñez-Sánchez, M.A.; Dávalos, A.; González-Sarrias, A.; Casas-Agustench, P.; Visioli, F.; Monedero-Saiz, T.; García-Talavera, N.V.; Gómez-Sánchez, M.B.; Sánchez-Álvarez, C.; García-Albert, A.M.; et al. MicroRNAs expression in normal and malignant colon tissues as biomarkers of colorectal cancer and in response to pomegranate extracts consumption: Critical issues to discern between modulatory effects and potential artefacts. Mol. Nutr. Food Res. 2015, 59, 1973–1986. [Google Scholar] [CrossRef]
- Ono, K.; Horie, T.; Nishino, T.; Baba, O.; Kuwabara, Y.; Kimura, T. MicroRNAs and High-Density Lipoprotein Cholesterol Metabolism. Int. Heart J. 2015, 56, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumitrescu, R.G. Early Epigenetic Markers for Precision Medicine. In Methods in Molecular Biology; Clifton, N.J., Ed.; Humana Press: New York, NY, USA, 2018; pp. 3–17. [Google Scholar]
- Kaur, S.; Abu-Shahba, A.G.; Paananen, R.O.; Hongisto, H.; Hiidenmaa, H.; Skottman, H.; Seppänen-Kaijansinkko, R.; Mannerström, B. Small non-coding RNA landscape of extracellular vesicles from human stem cells. Sci. Rep. 2018, 8, 15503. [Google Scholar] [CrossRef]
- Ioannidis, J.; Donadeu, F.X. Comprehensive analysis of blood cells and plasma identifies tissue-specific miRNAs as potential novel circulating biomarkers in cattle. BMC Genomics 2018, 19, 243. [Google Scholar] [CrossRef] [PubMed]
- Casas-Agustench, P.; Iglesias-Gutiérrez, E.; Dávalos, A. Mother’s nutritional miRNA legacy: Nutrition during pregnancy and its possible implications to develop cardiometabolic disease in later life. Pharmacol. Res. 2015, 100, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-alvarado, M.I.; Mercader, J.M.; Moreno-navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Molina-Torres, J.; Pérez-Luque, E.L.; et al. ScienceDirect Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Aganzo, M.; Montojo, M.-T.; López de Las Hazas, M.-C.; Martínez-Descals, A.; Ricote-Vila, M.; Sanz, R.; González-Peralta, I.; Martín-Hernández, R.; de Dios, O.; Garcés, C.; et al. Customized Dietary Intervention Avoids Unintentional Weight Loss and Modulates Circulating miRNAs Footprint in Huntington’s Disease. Mol. Nutr. Food Res. 2018, 62, e1800619. [Google Scholar] [CrossRef]
- Parnell, L.D.; Ordovas, J.M.; Lai, C.-Q. Environmental and epigenetic regulation of postprandial lipemia. Curr. Opin. Lipidol. 2018, 29, 30–35. [Google Scholar] [CrossRef]
- Bär, C.; Thum, T.; De Gonzalo-Calvo, D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics 2019, 11. [Google Scholar] [CrossRef]
- Mennigen, J.A.; Panserat, S.; Larquier, M.; Plagnes-Juan, E.; Medale, F.; Seiliez, I.; Skiba-Cassy, S. Postprandial Regulation of Hepatic MicroRNAs Predicted to Target the Insulin Pathway in Rainbow Trout. PLoS ONE 2012, 7, e38604. [Google Scholar] [CrossRef]
- Sun, C.; Alkhoury, K.; Wang, Y.I.; Foster, G.A.; Radecke, C.E.; Tam, K.; Edwards, C.M.; Facciotti, M.T.; Armstrong, E.J.; Knowlton, A.A.; et al. IRF-1 and miRNA126 Modulate VCAM-1 Expression in Response to a High Fat Meal. Circ. Res. 2012, 111, 1054–1064. [Google Scholar] [CrossRef]
- Lopez, S.; Bermudez, B.; Montserrat-de, S.; Abia, R.; Muriana, F.J. ScienceDirect high-saturated-fat challenge. J. Nutr. Biochem. 2018, 57, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Corraze, G.; Plagnes-Juan, E.; Quillet, E.; Dupont-Nivet, M.; Skiba-Cassy, S. Regulation of genes related to cholesterol metabolism in rainbow trout (Oncorhynchus mykiss) fed a plant-based diet. Am. J. Physiol. Integr. Comp. Physiol. 2018, 314, R58–R70. [Google Scholar] [CrossRef] [PubMed]
- Talbot, C.P.; Mensink, R.P.; Smolders, L.; Bakeroot, V.; Plat, J. Theobromine Does Not Affect Fasting and Postprandial HDL Cholesterol Efflux Capacity, While It Decreases Fasting miR-92a Levels in Humans. Mol. Nutr. Food Res. 2018, 62, 1800027. [Google Scholar] [CrossRef] [PubMed]
- Qiang, J.; Tao, Y.F.; Bao, J.W.; Chen, D.J.; Li, H.X.; He, J.; Xu, P. High Fat Diet-Induced miR-122 Regulates Lipid Metabolism and Fat Deposition in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus) Liver. Front. Physiol. 2018, 9, 1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Zamorano, J.; Martin-Hernandez, R.; Daimiel, L.; Richardson, K.; Giordano, E.; Nicod, N.; García-Carrasco, B.; Soares, S.M.A.; Iglesias-Gutiérrez, E.; Lasunción, M.A.; et al. Docosahexaenoic Acid Modulates the Enterocyte Caco-2 Cell Expression of MicroRNAs Involved in Lipid Metabolism. J. Nutr. 2014, 144, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.W.; Bass, B.L. A Role for the RNase III Enzyme DCR-1 in RNA Interference and Germ Line Development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating miRNAs Regulate Gene Expression in Other Tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Rotllan, N.; Price, N.; Pati, P.; Goedeke, L.; Fernández-Hernando, C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis 2016, 246, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Chomzynski, P.; Sacchi, N. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate–Phenol–Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Hyson, D.; Rutledge, J.C.; Berglund, L. Postprandial lipemia and cardiovascular disease. Curr. Atheroscler. Rep. 2003, 5, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Dávalos, A.; Fernández-Hernando, C. From evolution to revolution: miRNAs as pharmacological targets for modulating cholesterol efflux and reverse cholesterol transport. Pharmacol. Res. 2013, 75, 60–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briand, O.; Touche, V.; Colin, S.; Brufau, G.; Davalos, A.; Schonewille, M.; Bovenga, F.; Carrière, V.; De Boer, J.F.; Dugardin, C.; et al. Liver X Receptor Regulates Triglyceride Absorption Through Intestinal Down-regulation of Scavenger Receptor Class B, Type 1. Gastroenterology 2016, 150, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, L.; Li, J.; Sun, P.; Shang, L.; Zhang, J.; Zhao, Q.; Ouyang, Y.; Li, L.; Gong, K. MicroRNA profiling of diabetic atherosclerosis in a rat model. Eur. J. Med Res. 2018, 23, 55. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Ma, F.; Li, W.; Ouyang, S.; Liu, Z.; Wu, J. miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway. Int. J. Mol. Sci. 2017, 18, 1510. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.W.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirby, T.J.; Chaillou, T.; McCarthy, J.J. The role of microRNAs in skeletal muscle health and disease. Front. Biosci. 2015, 20, 37–77. [Google Scholar] [Green Version]
- Siracusa, J.; Koulmann, N.; Sourdrille, A.; Chapus, C.; Verret, C.; Bourdon, S.; Goriot, M.-E.; Banzet, S. Phenotype-Specific Response of Circulating miRNAs Provides New Biomarkers of Slow or Fast Muscle Damage. Front. Physiol. 2018, 9, 684. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.; Qin, X.; Luo, X.; Zhang, Y.; Liu, Z.; Zhu, L. Identification of miRNAs associated with the mechanical response of hepatic stellate cells by miRNA microarray analysis. Exp. Ther. Med. 2018, 16, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Rau, C.-S.; Jeng, J.C.; Chen, Y.-C.; Lu, T.-H.; Wu, C.-J.; Wu, Y.-C.; Tzeng, S.-L.; Yang, J.C.-S.; Yang, J.C. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J. Biomed. Sci. 2012, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- McKenna, L.B.; Schug, J.; Vourekas, A.; McKenna, J.B.; Bramswig, N.C.; Friedman, J.R.; Kaestner, K.H. MicroRNAs Control Intestinal Epithelial Differentiation, Architecture, and Barrier Function. Gastroenterol. 2010, 139, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-C.; Sahasrabuddhe, N.A.; Kim, M.-S.; Getnet, D.; Yang, Y.; Peterson, J.M.; Ghosh, B.; Chaerkady, R.; Leach, S.D.; Marchionni, L.; et al. Regulation of Lipid Metabolism by Dicer Revealed through SILAC Mice. J. Proteome Res. 2012, 11, 2193–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, R.A.; Deasy, W.; Hayes, A.; Cooke, M.B. High fat diet and associated changes in the expression of micro-RNAs in tissue: Lessons learned from animal studies. Mol. Nutr. Food Res. 2017, 61, 1600943. [Google Scholar] [CrossRef]
- Nezami, B.G.; Mwangi, S.M.; Lee, J.E.; Jeppsson, S.; Anitha, M.; Yarandi, S.S.; Farris, A.B., III; Srinivasan, S. MicroRNA 375 Mediates Palmitate-Induced Enteric Neuronal Damage and High-Fat Diet-Induced Delayed Intestinal Transit in Mice. Gastroenterology 2014, 146, 473–483. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Toledo, J.B.; Tsivinsky, V.G.; Irwin, D.; Grossman, M.; Weintraub, D.; Hurtig, H.I.; Chen-Plotkin, A.; Wolk, D.A.; McCluskey, L.F.; et al. Circulating brain-enriched microRNAs as novel biomarkers for detection and differentiation of neurodegenerative diseases. Alzheimer’s Res. Ther. 2017, 9, 89. [Google Scholar] [CrossRef]
- Li, D.-B.; Liu, J.-L.; Wang, W.; Luo, X.-M.; Zhou, X.; Li, J.-P.; Cao, X.-L.; Long, X.-H.; Chen, J.-G.; Qin, C. Plasma Exosomal miRNA-122-5p and miR-300-3p as Potential Markers for Transient Ischaemic Attack in Rats. Front. Aging Neurosci. 2018, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Morales, P.E.; Bucarey, J.L.; Espinosa, A. Muscle Lipid Metabolism: Role of Lipid Droplets and Perilipins. J. Diabetes Res. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Dong, K.; Xu, Y.; Yang, Q.; Shi, J.; Jiang, J.; Chen, Y.; Song, C.; Wang, K. Associations of Functional MicroRNA Binding Site Polymorphisms in IL23/Th17 Inflammatory Pathway Genes with Gastric Cancer Risk. Mediat. Inflamm. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Raju, G.S.; Chang, D.W.; Lin, S.-H.; Chen, Z.; Wu, X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer 2018, 124, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.; Di Bella, S.; Nigita, G.; Macca, V.; Lagana, A.; Giugno, R.; Pulvirenti, A.; Ferro, A. miRandola: Extracellular Circulating MicroRNAs Database. PLoS ONE 2012, 7, e47786. [Google Scholar] [CrossRef] [PubMed]
- Parizadeh, S.M.; Jafarzadeh-Esfehani, R.; Ghandehari, M.; Parizadeh, S.M.R.; Hassanian, S.M.; Rezayi, M.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. Circulating exosomes as potential biomarkers in cardiovascular disease. Curr. Pharm. Des. 2018, 25, 4436–4444. [Google Scholar] [CrossRef] [PubMed]
- Matheny, R.W.; Carrigan, C.T.; Abdalla, M.N.; Geddis, A.V.; Leandry, L.A.; Aguilar, C.A.; Hobbs, S.S.; Urso, M.L. RNA transcript expression of IGF-I/PI3K pathway components in regenerating skeletal muscle is sensitive to initial injury intensity. Growth Horm. IGF Res. 2017, 32, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastorakos, G.; Zapanti, E. The Hypothalamic-Pituitary-Adrenal Axis in the Neuroendocrine Regulation of Food Intake and Obesity: The Role of Corticotropin Releasing Hormone. Nutr. Neurosci. 2004, 7, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Singh, A.K.; Rotllan, N.; Goedeke, L.; Wing, A.; Canfrán-Duque, A.; Diaz-Ruiz, A.; Araldi, E.; Baldán, Á.; Camporez, J.-P.; et al. Genetic Ablation of miR-33 Increases Food Intake, Enhances Adipose Tissue Expansion, and Promotes Obesity and Insulin Resistance. Cell Rep. 2018, 22, 2133–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.-S.; Lin, Y.; Sun, F.-B.; Gao, J.; Han, B.; Li, S.J. miR-409 down-regulates Jak-Stat pathway to inhibit progression of liver cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 146–154. [Google Scholar] [PubMed]
- Winter, J. MicroRNAs of the miR379–410 cluster: New players in embryonic neurogenesis and regulators of neuronal function. Neurogenesis 2015, 2, e1004970. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Fernández-Hernando, C. miRNA regulation of white and brown adipose tissue differentiation and function. Biochim. Biophys. Acta 2016, 1861, 2104–2110. [Google Scholar] [CrossRef] [Green Version]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [Green Version]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, B.T.; Zhu, L.; Eckel, R.H.; Stafford, J.M. Sex differences in lipid and lipoprotein metabolism. Mol. Metab. 2018, 15, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.M.U.P.; Lopes, L.L.; Da Silva, A.; Oliveira, L.L.; Bressan, J.; Hermsdorff, H.H.M. Orange juice modulates proinflammatory cytokines after high-fat saturated meal consumption. Food Funct. 2017, 8, 4396–4403. [Google Scholar] [CrossRef] [PubMed]






| miRNA | p-Value | miRNA | p-Value |
|---|---|---|---|
| miR-466b-5p | 1.36 × 10−5 | miR-208a-5p | 0.403192 |
| miR-206-3p | 9.30 × 10−5 | miR-137-3p | 0.407196 |
| miR-10a-3p | 0.000143 | miR-127-5p | 0.415694 |
| miR-1198-3p | 0.000287 | miR-762 | 0.44027 |
| miR-543-3p | 0.000857 | miR-489-3p | 0.443628 |
| miR-450a-2-3p | 0.000977 | miR-470-3p | 0.48737 |
| miR-466c-5p | 0.001046 | miR-20a-3p | 0.510021 |
| miR-215-5p | 0.003314 | miR-700-3p | 0,524019 |
| miR-27b-5p | 0.003793 | miR-34b-5p | 0.541163 |
| miR-409-3p | 0.006274 | miR-291a-3p | 0.565681 |
| miR-1982-5p | 0.01161 | miR-153-3p | 0.585034 |
| miR-496a-3p | 0.013289 | miR-467c-5p | 0.586 |
| miR-340-3p | 0.027317 | miR-671-3p | 0.596674 |
| miR-183-3p | 0.030129 | miR-291a-5p | 0.597004 |
| miR-1941-3p | 0.03114 | miR-615-3p | 0.602924 |
| miR-130b-5p | 0.041939 | miR-376c-5p | 0.619959 |
| miR-125-3p | 0.050039 | miR-182-5p | 0.623672 |
| miR-1251 | 0.053334 | miR-1b-5p | 0.62594 |
| miR-542-3p | 0.063337 | miR-299a-3p | 0.629587 |
| miR-10b-3p | 0.147649 | miR-490-5p | 0.630007 |
| miR-1943-5p | 0.201154 | miR-1927 | 0.630042 |
| miR-329-3p | 0.21081 | miR-380-3p | 0.63362 |
| miR-20b-3p | 0.211707 | miR1894-3p | 0.656185 |
| miR-680 | 0.217664 | miR-342-5p | 0.668416 |
| miR-331-5p | 0.288161 | miR-342-5p | 0.678952 |
| miR-1186a | 0.29046 | miR-325-3p | 0.693872 |
| miR-335-3p | 0.294498 | miR-21a-3p | 0.704326 |
| miR-804 | 0.31838 | miR-743b-5p | 0.724951 |
| miR-667-3p | 0.325072 | miR-216a-5p | 0.758007 |
| miR-468-3p | 0.371391 | miR-323-3p | 0.774188 |
| miR-291b-5p | 0.373164 | miR-129-5p | 0.791411 |
| miR-295-5p | 0.390927 | miR-1953 | 0.834637 |
| miR-194-2-3p | 0.396934 | miR-207 | 0.892798 |
| miR-875-5p | 0.401872 | miR-6691-5p | 0.92054 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mantilla-Escalante, D.C.; López de las Hazas, M.-C.; Gil-Zamorano, J.; del Pozo-Acebo, L.; Crespo, M.C.; Martín-Hernández, R.; del Saz, A.; Tomé-Carneiro, J.; Cardona, F.; Cornejo-Pareja, I.; et al. Postprandial Circulating miRNAs in Response to a Dietary Fat Challenge. Nutrients 2019, 11, 1326. https://doi.org/10.3390/nu11061326
Mantilla-Escalante DC, López de las Hazas M-C, Gil-Zamorano J, del Pozo-Acebo L, Crespo MC, Martín-Hernández R, del Saz A, Tomé-Carneiro J, Cardona F, Cornejo-Pareja I, et al. Postprandial Circulating miRNAs in Response to a Dietary Fat Challenge. Nutrients. 2019; 11(6):1326. https://doi.org/10.3390/nu11061326
Chicago/Turabian StyleMantilla-Escalante, Diana C., María-Carmen López de las Hazas, Judit Gil-Zamorano, Lorena del Pozo-Acebo, M. Carmen Crespo, Roberto Martín-Hernández, Andrea del Saz, Joao Tomé-Carneiro, Fernando Cardona, Isabel Cornejo-Pareja, and et al. 2019. "Postprandial Circulating miRNAs in Response to a Dietary Fat Challenge" Nutrients 11, no. 6: 1326. https://doi.org/10.3390/nu11061326