Published online Jun 28, 2014. doi: 10.3748/wjg.v20.i24.7830
Revised: February 7, 2014
Accepted: March 12, 2014
Published online: June 28, 2014
An improvement in pancreatic cancer treatment represents an urgent medical goal. Late diagnosis and high intrinsic resistance to conventional chemotherapy has led to a dismal overall prognosis that has remained unchanged during the past decades. Increasing knowledge about the molecular pathogenesis of the disease has shown that genetic alterations, such as mutations of K-ras, and especially epigenetic dysregulation of tumor-associated genes, such as silencing of the tumor suppressor p16ink4a, are hallmarks of pancreatic cancer. Here, we describe genes that are commonly affected by epigenetic dysregulation in pancreatic cancer via DNA methylation, histone acetylation or miRNA (microRNA) expression, and review the implications on pancreatic cancer biology such as epithelial-mesenchymal transition, morphological pattern formation, or cancer stem cell regulation during carcinogenesis from PanIN (pancreatic intraepithelial lesions) to invasive cancer and resistance development. Epigenetic drugs, such as DNA methyltransferases or histone deactylase inhibitors, have shown promising preclinical results in pancreatic cancer and are currently in early phases of clinical development. Combinations of epigenetic drugs with established cytotoxic drugs or targeted therapies are promising approaches to improve the poor response and survival rate of pancreatic cancer patients.
Core tip: Pancreatic cancer represents a devastating disease with poor overall survival at advanced stages, and new and effective treatment options are required. Besides genetic mutations, epigenetic dysregulation of oncogenes and tumor suppressor genes is recognized as a novel therapeutic target. Mechanisms underlying DNA methylation, histone acetylation and microRNA regulation and their contribution to pancreatic cancer development and resistance to treatment are highlighted in this review. Potential therapeutic interventions are discussed.
- Citation: Neureiter D, Jäger T, Ocker M, Kiesslich T. Epigenetics and pancreatic cancer: Pathophysiology and novel treatment aspects. World J Gastroenterol 2014; 20(24): 7830-7848
- URL: https://www.wjgnet.com/1007-9327/full/v20/i24/7830.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i24.7830
Overall, the incidence of pancreatic cancer is minimally increasing or stable[1]. Experimental and clinical investigations have been intensified in the last few years (1) to obtain more pathogenetic insights in this highly life-destructive cancer entity; (2) to improve the early detection rate of this often concealed disease; and (3) to identify new therapeutic strategies to improve quality of life and survival time[2,3]. Nevertheless, the fate of patients with a diagnosis of pancreatic cancer is miserable even with maximal application of possible combined therapeutic interventions such as surgery, radiation and/or chemotherapy[4,5]. The overall survival time of patients with pancreatic cancer is a mean of 1 year after diagnosis[6]. This leads to the unsettling question of whether patients with a diagnosis of pancreatic cancer can survive at all[7].
In the last few years, one therapeutic point of attack has been concerned with the role of cancer stem-cells and the epithelial-mesenchymal transition under the influence of epigenetic regulator mechanisms[8]. These approaches are interesting and promising as they could explain the chemotherapy refractiveness of most pancreatic cancers. We have shown previously that pancreatic cancer employs classical pathways of organ development and embryogenesis such as Hedgehog or WNT (wingless) signaling[9,10] which, amongst others, could be targets for novel therapeutic approaches.
In this review, we focus on epigenetic regulation mechanisms in pancreatic cancer providing possible novel treatment aspects by highlighting the pathophysiology of this special tumor entity for pathologists, clinicians and future therapeutic approaches.
Pancreatic cancer is associated with a high mortality rate and represents the 7th most frequent cause of cancer death, with approximately 265000 deaths and an incidence of 280000 per year worldwide in 2008[11,12]. Europe and Northern America have the highest incidence of pancreatic cancer, with slightly more males being affected.
Pancreatic cancer is usually diagnosed at an advanced stage due to a lack of symptoms in the early stages so that resection of the advanced tumor is often not possible. The overall 1-year survival rate for pancreatic cancer is 26%, and the 5-year survival rate is approximately 6% for advanced cancer and 22% for early stages when surgical removal of the tumor is still possible[13].
Therefore, new therapeutic approaches combining neoadjuvant chemotherapy and radiotherapy to significantly reduce the tumor size are promising to allow the option of surgical removal in selected patients[14].
Classical malignant pancreatic tumors show heterogeneous glandular and duct-like, grading-dependent structures, mostly infiltrating the pancreatic parenchyma and exhibiting a partially prominent desmoplastic stroma. The widely accepted and routinely used grading system of pancreatic cancer is based on (1) glandular differentiation; (2) mucin production; (3) mitosis (per 10 microscopic high power fields), and (4) nuclear features (such as nuclear polymorphism, size or arrangement of the nucleus)[15,16]. So far, no definitive and routinely used immunohistochemical markers exist, although many biological markers in pancreatic ductal adenocarcinoma were tested as possible diagnostic and prognostic tools. However, the main limitations arise from the small number of patients studied and in the heterogeneity of the applied methods[17]. Further approaches for prognostic grading focused on different morphological patterns similar to Gleason’s scoring system[18], included epithelial-mesenchymal characteristics, such as vimentin expression and tumor budding[19,20], or evaluated several gene expression signatures including downregulation of ASPM (abnormal spindle-like microcephaly associated) which could be detected by immunohistochemistry[21].
Detailed morphological analysis revealed prognostic subtypes of pancreatic cancer, with a group associated with better survival (colloid and medullary) and a group with a worse outcome (adenosquamous or undifferentiated)[22] (as described in Table 1).
| Variant (source) | Morphology | Genetics/epigenetics | Prognosis |
| MC | Well defined pushing border, syncytial growth pattern, and poorly differentiated cancer cells | Germline or somatic mutations as well as epigenetic silencing by promoter methylation of mismatch repair genes MLH1 and MSH2 | BetterMC: Overall 2- and 5-yr survival rate of 29% and 13%[23]CC: 2-yr and 5-yr survival rate of 70% and 57%[153] |
| CC | Suspension of well-differentiated cancer cells in extracellular mucin pools (at least 80%) | Unknown | |
| AC | Combination of glandular and squamous (at least 30%) components | K-RAS2 mutations, inactivation of CDKN2A/p16, SMAD4/DPC4 and TP53 | WorseAC/UC: Median survival of 5 mo after resection[154,155] |
| UC | Noncohesive cancer, lacking histological features of differentiation | K-RAS2 gene mutations, loss of E-cadherin protein expression (promoter hypermethylation) | |
| Expression of L1CAM, COX2, and EGFR | |||
| Subtype with osteoclast-like giant cells shows mutations like noninvasive precursor lesions |
It is important not only to give the diagnosis “pancreatic cancer”, but to discriminate between tumor entities for patient communication, and to evaluate a possible family history of cancer in genetic counselling (as in cases of medullary carcinoma[23]) as well as to establish tumor-specific therapy modalities, since it is possible to link tumor sub-entities to specific genetic lesions (Table 1).
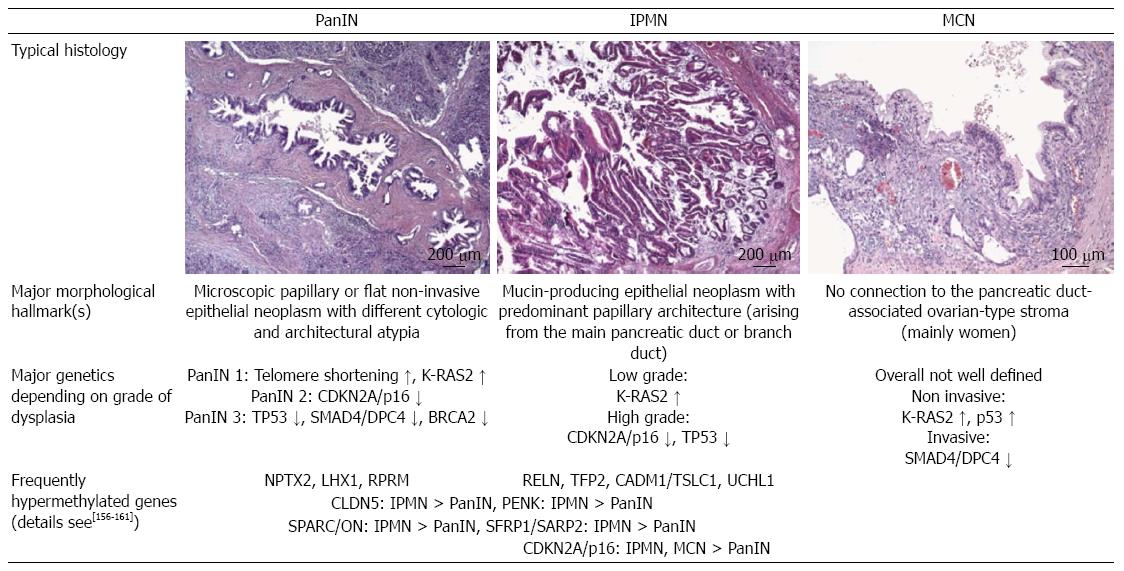
Pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasms (IPMN) and mucinous cystic neoplasms (MCN) are considered precursor lesions[24] (Figure 1). As IPMN and MCN are seen pre-therapeutically in radiological investigations, therapeutic strategies/algorithms were established to weigh up the extent of surgical resection and patients’ quality of life.
Further molecular analyses of different precursor lesions and their morphological variants revealed a stepwise model of carcinogenesis and possible epigenetic associations (Figure 1). Interestingly, a huge number of epigenetically regulated genes have been detected by global gene expression profiles in comparison with classical genetic alterations showing an association with cytological and architectural atypia of these precursor lesions. The challenge in the future is to analyze the complex mechanistic crosstalk of these genetic and epigenetic regulatory mechanisms[25] during pancreatic carcinogenesis for new drug development and administration, as shown by Breitkreutz et al[26] using network statistics.
Intensive DNA analysis using genome- and epigenome-wide screening methods during the last few years have elucidated some major deregulated gate “drivers”, “passengers” and “keepers” in pancreatic cancer (Table 2)[27-30]. Overall, more suppressor genes than oncogenes are involved in pancreatic cancer.
| Gene symbol (source) | Associated function according the hallmarks of cancer1[35,36] | Frequency | Type of genetic alteration | Evidence for epigenetic regulation (reference) |
| Activation | ||||
| K-RAS2[131] | a, f, g | > 90 | Point mutation | |
| AKT2[162,163] | a, j, g | 10-20 | Amplification | |
| BRAF[164] | a, b, g | 5 | Point mutation | |
| Deactivation | ||||
| CDKN2A/p16[165] | b, d, i | 95 | Homozygous deletion, intragenic mutation | Yes[166] |
| TP53[167-169] | b, d, i, h | 50-70 | Intragenic mutation an one allele and loss in the other allele | |
| SMAD4/DPC4[170,171] | b, c, f | 55 | Homozygous deletion, intragenic mutation | |
| MLH1[23,172] | h | 3-15 | Heterozygote mutations | Yes[34] |
| BRCA2[173] | a | 7 | Heterozygote mutations | |
| STK11/LKB1[174] | i | 5 | Homozygous deletion, intragenic mutation | |
| TGFBR2[175] | a, f | 4 | Homozygous deletion, homozygous frameshift mutation | |
| MAP2K4[176,177] | a | 2 | Homozygous deletion, missense mutation | |
As recently described in depth by Hong et al[31], the most frequently mutated oncogene (> 95%) in pancreatic cancer is K-RAS (Kirsten rat sarcoma viral oncogene homolog), leading to constitutive downstream signaling of proliferation, cellular survival, motility and remodeling. On the other hand, the major deregulated suppressor genes in pancreatic cancer are CDKN2A/p16 (cyclin-dependent kinase inhibitor 2A), TP53 (tumor protein p53) and SMAD4/DPC4 (SMAD family member 4) which are inactivated by 2 different, often independent, mechanisms. Whereas CDKN2A/p16 and TP53 are mainly involved in cell cycle checkpoint control and arrest, SMAD4/DPC4 plays an important role in signal transduction of the transforming growth factor (TGF)-β pathway, and furthermore in cellular proliferation. Finally, when looking at epigenetically affected genes, it is obvious that the classical and most frequent pancreatic cancer genes are only partially epigenetically regulated (Table 2). Interestingly, such cases with epigenetically silenced MLH1 (mutL homolog 1) genes are associated with the distinctive medullary phenotype of pancreatic cancer[32-34].
As later described in detail (Figure 1, Tables 3 and 4), epigenetics via DNA methylation, histone acetylation or interacting regulative microRNAs (miRNAs) could essentially be linked to different morphological and genetic changes during pancreatic carcinogenesis. Extensive investigations are been carried out on epigenetic changes in pancreatic cancer precursor lesions, indicating that heterogeneous, non-linked pathways of carcinogenesis are regulated by epigenetics as summarized in Figure 1 and Table 3. Detailed analysis of the function of these epigenetically deregulated genes revealed that all hallmarks of cancer[35,36] such as self-sufficiency in growth signals (e.g., SFRP1/SARP2; secreted frizzled-related protein 1), insensitivity to anti-growth signals [e.g., CDKN2A/p16 or RPRM (reprimo)], tissue invasion and metastasis [e.g., SPARC/ON (secreted protein, acidic, cysteine-rich (osteonectin)], limitless replicative potential [e.g., LHX1 (LIM homeobox 1)], sustained angiogenesis [e.g., CLDN5 (Claudin-5)] or evading apoptosis [e.g., RPRM or CADM1/TSLC1 (cell adhesion molecule 1/tumor suppressor in lung cancer 1)] are involved in pancreatic cancer and affected by epigenetic (de)regulation. This supports our knowledge of the pleiotropic effects of systemic epigenetic mechanisms. The degree of cytological and architectural atypia correlated with the amount of methylated genes supporting the hypothesized multiple step model of pancreatic cancer even in the early disease stages[31,37].
| DNA modification | Material | Gene affected | Ref. | ||
| Cancer samples | Cell lines | Other | |||
| DNA hyper-methylation | √ | p16 | [166] | ||
| √ | RASSF1A | [178] | |||
| √ | MDFI, hsa-miR-9-1, ZNF415, CNTNAP2, ELOVL4 | [179] | |||
| √ | SOX15 | [180] | |||
| √ | HOP hoemobox (HOPX) | [181] | |||
| √ | KLF10 | [182] | |||
| √ | hMLH1 | [183] | |||
| √ | miR-34a/b/c | [184] | |||
| √ | SPARC | [185] | |||
| √ | FoxE1, NPTX2, CLDN5, P16, TFPI-2, SPARC, ppENK | [186] | |||
| √ | SFRP | [187] | |||
| √ | AsPC1, Hs766T, MiaPaCa2, Panc1 | UCHL1, NPTX2, SARP2, CLDN5, reprimo, LHX1, WNT7A, FOXE1, TJP2, CDH3, ST14 | [30,159] | ||
| √ | AsPC1, BxPC3, CFPAC1, Panc1 | NPTX2 | [188] | ||
| √ | Panc1, SW1990 | miR-132 | [121] | ||
| √ | BxPC3, Capan2, CFPAC1, HPACI, HPAFII, MiaPaCa2, Panc1, PL45 | FOXA1/2 | [81] | ||
| MiaPaCa2 | ARID1B | [189]1 | |||
| Panc1 | NPTX2 | [190]1 | |||
| AsPC1, BxPC3, Panc1, MIA PaCa-2 | Dkk3 | [191] | |||
| BXPC3, HPAFII, HPAC, hTERT-HPDE, Panc1 | Cldn18 | [192] | |||
| BxPC3, CFPAC1, Panc1, SW1990 | TNFRSF10C | [193] | |||
| Pancreatic juice | Neuronal pentraxin II (NPTX2) | [194] | |||
| Pancreatobiliary fluid | UCHL1, RUNX3 | [195] | |||
| PanIN | p16 | [196] | |||
| IPMNs | BNIP3, PTCHD2, SOX17, NXPH1, EBF3, SPARC, SARP2, TSLC1, RELN, TFPI2, CLDN5, UCHL1 | [157,197] | |||
| Blood, brush cytology | NPTX2 | [198,199] | |||
| DNA hypo-methylation | √ | VAV1 | [79] | ||
| √ | Claudin4, lipocalin2, 14-3-3 sigma, trefoil factor 2, S100A4, mesothelin, prostate stem cell antigen | [78] | |||
| √ | MUC4 | [77] | |||
| SW1990 | ABCB1/MDR1, ABCC1/MRP1, ABCG2/BCRP | [200] | |||
| miRNA/function | Cell lines | Target gene(s) | Cellular effects | Ref. | |
| Function as oncogene | -10a | AsPC1, Capan1, Capan2, MiaPaCa2, Panc1, Patu8988T, Patu8988S, Patu8902 | HOXB1, 3 | Metastasis ↑ | [201] |
| -21 | AsPC1, BxPC3, Capan1, Capan2, CFPAC1, Hs776T, H48N, KP-1N, KP-2, KP-3, MiaPaCa2, NOR-P1, Panc1, SUIT-2, SW1990 | HOXA1 | Invasion ↑ | [202] | |
| AsPC1, Capan1, Capan2, CFPAC1, H48N, HS766T, KP-1N, KP-2, KP-3, MiaPaCa2, NOR-P1, Panc1, SUIT-2, SW1990 | Proliferation ↑, invasion ↑, chemoresistance ↑ | [203] | |||
| BxPC3 | Proliferation ↑ | [204] | |||
| Capan1, HS766T, MiaPaCa2, MPanc96, Panc1, PL45, SW1990 | PTEN, RECK | After miRNA inhibition: cell cycle arrest ↑, apoptosis ↑ | [205] | ||
| -132, -212 | Panc1 | Rb1 | Proliferation ↑ | [206] | |
| -155 | Capan2, MiaPaCa2, MCF7, MEFs, 293T | TP53INP1 | Apoptosis ↓ | [207] | |
| -194, -200b, -200c, -429 | AsPC1, A818, BxPC3, Capan1, Capan2, HPAFII, MiaPaCa2, MPanc96, Panc1, Patu8902, Patu8988T, Patu8988S, PT45, Suit 007, Su.86.86, Sut00281 | EP300 | Metastasis ↑ | [208] | |
| -197 | AsPC1, Panc1 | p120 catenin | EMT ↑ | [209] | |
| -210 | Panc1, MiaPaCa2, SUIT-2 | Migration ↓, vimentin ↓, snai-1 ↓, membraneous β-catenin ↑ | [210] | ||
| -221 | Capan1, HS766T, MiaPaCa2, MPanc96, Panc1, PL45, SW1990 | p27 | Chemosensitivity ↑ | [205] | |
| -224, -486 | AsPC1, A818, BxPC3, Capan1, Capan2, HPAFII, Su 86.86, MPanc96, MiaPaCa2, Panc1, Patu8902, Patu8988T, PT45, Patu8988S, Suit 007, Suit 00281 | CD40 | Invasion ↑, metastasis ↑ | [211] | |
| Function as tumor suppressor | -301a | BxPC3, Hs766T | Bim | Proliferation ↑, | [212] |
| -320c | AsPC1, Panc12 | SMARCC1 | Chemoresistance ↑ | [213] | |
| -421 | SW1990, Panc1 | DPC4/Smad4 | Proliferation ↑, colony formation ↑ | [214] | |
| -491-5p | AsPC1, Capan1, MiaPaCa2, SW1990 | Bcl-XL, TP53 | Proliferation ↓, apoptosis ↑, STAT3 ↓, PI-3K/Akt ↓ | [215] | |
| let-7let-7a | BxPC3, Capan1, Capan2, human HPNE (human pancreatic nestin-positive) cells MiaPaCa2, Panc1 | Proliferation ↓, K-RAS ↓, MAPK ↓ | [216] | ||
| AsPC1 | RAS | K-RAS ↓, radiosensitivity ↑ | [217] | ||
| -22 | BxPC3 | SP1, ESR1 | Tumourigenesis ↓ | [218] | |
| -26a | SW1990, Panc1 | HMGA1 | Proliferation ↓, invasion ↓, migration ↓, apoptosis ↑ | [219] | |
| -34 | BxPC3, MiaPaCa2 | Bcl-2, Notch-1/2 | Clonogenicity ↓, invasion ↓, apoptosis ↑, cell cycle arrest ↑, chemosensitivity ↑, radiosensitivity ↑, CSC ↓ | [220] | |
| -34a | Panc1 | Cell cycle arrest ↑, apoptosis ↑, migration ↓, E2F3 ↓, Bcl-2 ↓, c-myc ↓, cyclin D1 ↓ | [221] | ||
| -34b | AsPC1, MiaPaCa2 | Notch-1 | Proliferation, apoptosis | [222] | |
| Smad3 | Progression in vivo↑ | [223]3 | |||
| -107 | MiaPaCa2, Panc1 | CDK6 | Proliferation ↓ | [224] | |
| -126 | AsPC1, BxPC3, KLM-1, MiaPaCa2, Panc1 | ADAM9 | Migration ↓, invasion ↓, E-cadherin ↑ | [225] | |
| -132 | BxPC3, HPAFII, HPAC, Panc1 | Proliferation ↓, colony formation ↓, Akt ↓ | [121] | ||
| -143 | AsPC1, BxPC3, Capan2, HPAFII, MiaPaCa2, Panc1 | COX-2 | Proliferation ↓, MEK/MAPK ↓ | [226] | |
| Panc1 | ARHGEF1 (GEF1), ARHGEF2 (GEF2), K-RAS | Migration ↓, invasion ↓, metastasis ↓, E-cadherin ↑ | [227] | ||
| -148a | IMIM-PC2 | CDC25B | Proliferation ↓, colony formation ↓ | [228] | |
| -148b | AsPC1, BxPC3, MiaPaCa2, Panc1, SW1990 | AMPKα1 | Proliferation ↓, apoptosis ↑, cell cycle arrest ↑, invasion ↓, chemosensitivity ↑, tumourigenicity ↓ | [229] | |
| -150 | Colo357, HPAF, Panc10.05 | MUC4 | Proliferation ↓, clonogenicity ↓, migration ↓, invasion ↓, cellular adhesion ↑ | [230] | |
| -200 | AsPC1, BxPC3, Colo357, HPAC, MiaPaCa2, L3.6pl, Panc12 | EMT ↓ (ZEB1 ↓, slug ↓, vimentin ↓) | [231] | ||
| -375 | Proliferation ↓, cell cycle arrest ↑, apoptosis ↑ | [232]3 | |||
| -548d | Panc1 | Proliferation ↓, apoptosis ↑, cell cycle arrest ↑ | [233] | ||
Additionally, hypomethylation is also recognized in pancreatic cancer, leading to genomic instability by overexpression of genes and proteins in contrast to hypermethylation by silencing genes and subsequent protein expression. Serial genes like SERPINB5 (serpin peptidase inhibitor, clade B, member 5), CLDN4, stratifin, lipocalin-2, trefoil factor2, S100P (S100 calcium-binding protein P), mesothelin or prostate stem cell antigen are hypomethylated which causes uncontrolled or “dys”-regulated cell cycle progression, proliferation, differentiation or adhesion[31].
The identification of DNA methylation, histone modification and the action of miRNAs has profoundly increased the knowledge about the regulation of gene activity. Epigenetics studies the stable and inheritable patterns of altered gene expression independent of the primary DNA sequence[38], and has shown that dynamic traits of chromatin, reversible covalent modification of DNA, and post-transcriptional regulation centrally impact on gene expression and phenotypic characteristics[8,39]. With increasing evidence that tumorigenesis-associated cellular changes are caused by epigenetic alterations, the field of cancer research has evolved to incorporate oncogenic mechanisms beyond DNA mutations. Epigenetic mechanisms (see Table 5 for an overview about the major epigenetic mechanisms) are generally reversible. Together with the fact that epigenetic alterations may be even more prevalent than genetic aberrations, this is highly attractive in the conceptual approach of selecting and exploiting potential molecular targets for novel cancer therapeutics[8,40].
| Mechanism | Enzyme | Subclasses/components | Effect on target gene expression |
| DNA (de-) methylation | DNMT | DNMT1 (methylation maintenance) | ↓ |
| DNMT3A and -3B (de novo methylation) | |||
| DNA de-methylase | Not known | ↑ | |
| Histone (de-) acetylation | HAT | e.g., CBP, p300 | ↑ |
| Histone methylation | HDAC | Class I (HDACs-1-3, -8), class IIa (HDACs-4, -5, -7, -9), class IIb (HDACs-6, -10), class III (SIRT1-7), class IV (HDAC-11) | ↑ |
| PcG→ H3-K27-me3 | PRC1: CBX-2, 4, or 9, PHC-1, 2, or 3, BMI1, RING1A/B or RNF2 | ↓ | |
| → H3-K27-me3 maintenance | |||
| PRC2: EZH2, SUZ12, EED | |||
| →de novo H3-K27-me3 maintenance | |||
| TrxG → H3-K4-me3 | Several members | ↑ | |
| Post-transcriptional | miRNAs | 2578 mature miRNA (miRBase v20) | ↓ |
The methylation of DNA and subsequent silencing of a gene is catalyzed by DNA methyltransferases (DNMTs) which add a methyl group to the 5’ carbon of the cytosine pyrimidine ring. This occurs preferably in regions containing cytosine-guanine dinucleotides (CpGs); these CpG islands are preferentially located in regions corresponding to regulatory regions of many genes[41]. While DNMT1 is responsible for maintenance of parental DNA methylation patterns following replication, de novo DNA methylation is catalyzed by DNMT3A and DNMT3B enzymes[42]. The identification of DNA demethylases which remove the methyl group and reverse the action of DNMTs still warrants further research. DNA methylation was the first type of epigenetic alteration identified as responsible for inactivation of a tumor suppressor gene[43], and it is suggested that 100-400 hypermethylated CpG islands may exist in a given tumor[44].
Compared with DNA-based epigenetics, alterations in DNA-associated histones offer a greater variety of covalent epigenetic modifications, including phosphorylation, methylation, acetylation, ubiquitination and sumoylation, all with different degrees of modification (e.g., mono-, di-, and trimethylation)[45,46]. The nucleosome as the core building block of eukaryotic chromatin comprises histone octamers (dimers of H2A, H2B, H3 and H4, i.e., the nucleosome core particle, NCP) and about 146 base pairs of DNA[47]. Modifications to histones determine the packing of DNA into either tight and transcriptionally silent heterochromatin or transcriptionally active and open-structured euchromatin. These modifications affecting chromatin mobility and stability have been termed “marks” whose type, position, and combinations determine whether a given gene is expressed or silenced, i.e., the histone code[46,48]. This hypothesis postulates that local histone modifications determine the configuration of chromatin, alter the binding affinities of non-histone proteins, and subsequently influence the chromatin structure and accessibility of DNA for transcription[39,49]. Histone acetylation is thought to activate gene transcription and is catalyzed by histone acetyltransferases (HATs) which transfer an acetyl group from acetyl coenzyme A to the ε-amino group of lysine such as CREBBP (cAMP response element-binding protein binding protein), p300 and p300-CBP-associated factor (P/CAF). The reverse reaction is mediated by histone deacetylases (HDACs) which comprise a group of 18 isoenzymes identified to date. HDACs are classified into four groups (I-IV) based on their homology to yeast HDACs as summarized in Table 5[50,51].
The polycomb group proteins (PcG) repress gene activity by trimethylation of H3-K27 (histone 3 lysine 27) while TrxG proteins (Trithorax group) activate gene expression via H3-K4 histone trimethylation. The PcGs have 2 functions: polycomb repressive complex 1 (PRC1) maintains the silenced (H3-K27-me3, trimethylated) chromatin state and consists of CBX-2, 4, or 8 (chromobox homologue 2/4/8), PHC-1, 2, or 3 (polyhomeotic homologue 1/2/3), BMI1 (B-cell-specific Moloney murine leukemia virus integration site 1), and RING1A/B or RNF2 (RING finger domain protein). PRC2 initiates the repressive state by de novo trimethylation of H3-K27 and consists of EZH2 (enhancer of zeste homologue 2), SUZ12 (suppressor of zeste 12) and EED (embryonic ectoderm development)[39]. Together with other chromatin-modifying enzymes including DNMTs and HDACs, the initially constituted suppression via H3K27-3me by PRC2 is maintained by PRC1 and allows fine-tuned, context-dependent regulation of gene silencing.
miRNAs are short (18-25 nucleotides), phylogenetically conserved single-stranded RNA molecules without protein-coding functions involved in the silencing of messenger RNAs (mRNAs)[52,53]. This post-transcriptional repression is accomplished by (partial) base-pairing with the respective mRNA causing either (1) inhibition of translation initiation; (2) inhibition of translation elongation; or (3) mRNA decay initiated by deadenylation of the mRNA following recruitment of a deadenylase complex[54,55]. The functions added a new layer of regulatory mechanisms affecting virtually all cellular functions[56,57]. The interaction between miRNAs and their target mRNAs ultimately leads to reduced levels of the regulated mRNA/protein. The current release of miRBase (v20[58]) lists 2578 mature miRNA sequences and it is estimated that over 60% of human mRNAs are direct miRNA targets[59]. miRNAs can function as either suppressors or oncogenes and their regulatory importance in human tumorigenesis has been demonstrated for various cancer types[60,61]. While miRNAs are per se categorized as an epigenetic mechanism, their cancer-related expression itself may be subject to epigenetic regulation by the above-mentioned mechanisms of chromatin modulation[62,63].
In the following paragraphs, we focus on their role in tumorigenesis in pancreatic cancer including their potential therapeutic exploitation by “epidrugs”. For a general overview on cancer-related epigenetic mechanisms, we kindly refer the reader to recent comprehensive reviews: DNA methylation[64-66], histone (de)acetylation[67-70], histone methylation[71-73], and miRNAs[74-76].
DNA methylation in pancreatic cancer: As discussed in McCleary-Wheeler et al[8] DNA methylation may occur early during tumorigenesis of pancreatic cancer as some epigenetic alterations are already observed in the earliest lesion, i.e., PanIN-1A, and their importance may increase during further tumor progression[30]. In line with these findings, mucin 4 (MUC4) gene expression is increased from normal to precancerous lesions to pancreatic cancer associated with an increasing frequency of MUC4 promoter hypomethylation[77]. Furthermore, not only DNA methylation but also hypomethylation and thus aberrantly high expression of a gene may represent an epigenetic feature in pancreatic carcinoma[78,79]. Table 3 summarizes the currently available literature on (deregulated) DNA methylation in pancreatic cancer.
Aberrant activation of developmental signalling pathways such as Hedgehog represents a frequently observed trait in human cancers[9]. He et al[80] have shown that the Hedgehog transcription factor Gli1 (GLI family zinc finger 1) targets epigenetic modifiers in pancreatic cancer, namely DNMT1 and DNMT3a. After showing a concomitantly higher expression of Gli1 and the DNMTs in pancreatic tumor samples, the authors proved by pharmacological inhibition using cyclopamine, Gli1 overexpression and siRNA (small interfering RNA)-based Gli1 knockdown, that the DNMT proteins are positive targets of this oncogenic pathway, suggesting a cross-talk between aberrantly activated embryogenesis pathways and activation of oncogenic epigenetic mechanisms in pancreatic cancer.
Related to the important role of epithelial-mesenchymal transition (EMT) in pancreatic tumor progression, FOXA1 and FOXA2 (forkhead box A1/2) transcriptions factor were identified as effective antagonists of EMT in pancreatic cancer due to their ability to positively regulate E-cadherin expression: in a study by Song et al[81], FOXA1/2 expression was lost during malignant progression and their promoter was extensively hypermethylated in a pancreatic cancer cell line. As the demethylation-mediated reactivation of E-cadherin required concomitant FOXA2 expression, the authors concluded that suppression of FOXA1/2 is necessary and sufficient for EMT in the progression of pancreatic cancer[81].
From the data summarized in Table 3 and the mentioned examples, it is clear that numerous genes and signaling targets are epigenetically regulated by means of DNA methylation in pancreatic cancer. Importantly, not only does late-stage, malignant pancreatic cancer display aberrant DNA methylation, but also earlier, pre-malignant lesions. It is important to note that DNA methylation may act in cooperation with other epigenetic mechanisms (histone modification) to achieve stable silencing of, for example, individual tumor suppressor genes. In a series of studies, Yamada et al[82-84] have shown that different mucin variants are regulated by different and complementary epigenetic mechanisms: MUC1, MUC2 and MUC5AC by DNA methylation and H3-K9 histone methylation. This fact has to be considered in therapeutic approaches which aim at reversing deregulated DNA methylation patterns, for example.
Histone-based epigenetics in pancreatic cancer: Acetylation and methylation of histones represent the 2 epigenetic mechanisms based on histone modifications for which currently data exist in the context of pancreatic tumorigenesis. Several studies have demonstrated the general relevance of HDAC enzymes in pancreatic cancer by showing that (1) HDAC2 is increased in pancreatic ductal adenocarcinoma, especially in poorly differentiated tumors[85]; and (2) the expression of HDAC7 is significantly increased in pancreatic adenocarcinoma samples; HDAC7 expression furthermore allows for discrimination between pancreatic adenocarcinoma from other pancreatic tumors (serous cystadenoma, IMPN)[86]. The potential of therapeutic targeting of HDACs in pancreatic cancers has been reviewed by Schneider et al[87]. As discussed in this section, several studies have unequivocally demonstrated the relevance of histone-based epigenetic mechanisms including the (oncogenic) functions of PRC1/2 protein complexes to contribute to pancreatic tumorigenesis. These studies provided evidence of either altered/aberrant expression of these epigenetic regulators in pancreatic cancer samples or demonstrated the potential therapeutic exploitation of these mechanisms using cell-based in vitro studies.
An interesting cooperation between the ZEB1 (zinc finger E-box binding homeobox) transcription factor which is responsible for silencing of the E-cadherin gene (CDH1) and HDAC enzymes was investigated by Aghdassi et al[88]. In 25 surgical pancreatic cancer specimens, the authors found neither hypermethylation nor somatic mutations in the CDH1 gene, but complexes of ZEB1/HDAC attached to the CDH1 promoter. Knockdown of ZEB1 prevented this interaction resulting in histone acetylation and re-expression of E-cadherin. This study has provided additional insights into how EMT transcription factors cooperate with HDACs to silence E-cadherin, and thus promote EMT and tumor progression[89]. These data confirmed earlier results which demonstrated that downregulation of E-cadherin in metastatic pancreatic cancer cells is mediated by a repressor complex containing the EMT transcription factor Snail and the HDAC1 and HDAC2 enzymes[90].
The particular role of PRC proteins as epigenetic mechanisms has only recently become a focus in pancreatic cancer research. In several tumor types, overexpression of the EZH2 gene is associated with poor prognosis, advanced stage, invasion and metastasis - for an overview see Crea et al[91]. As reviewed in Grzenda et al[39], EZH2 overexpression promoted survival, angiogenesis, migration, proliferation and repression of E-cadherin. For pancreatic cancer, several studies demonstrated an involvement of components of PRC1/2 in malignant progression: Martínez-Romero et al[92] found the expression of the PRC1 proteins Bmi1 and Ring1b to be upregulated in PanIN lesions (Bmi1) and pancreatic adenocarcinoma (both components). These finding were confirmed by Song et al[93] showing that Bmi1 is correlated with lymph node metastasis and poor survival rates; furthermore, stable knockdown of Bmi1 reduced the levels of cyclin D1, CDK-2/4 (cyclin-dependent kinase), Bcl-2 (B-cell CLL/lymphoma 2) and phosphor-Akt while the expression of p21 and Bax was increased and associated with higher susceptibility towards apoptosis induction. Moreover, Wellner et al[94] showed that Bmi1 and other “stemness” factors were negative targets of the epithelial differentiation-linked miR-200c, -203 and -183 suggesting a possible mechanism for Bmi1 deregulation in pancreatic cancer. Another PRC1 member was investigated by Karamitopoulou et al[95]: CBX7 was analyzed in 210 ductal pancreatic adenocarcinomas and its expression was found to progressively decrease from normal pancreatic tissue, PanINs and invasive ductal adenocarcinoma-associated with increasing malignancy and a trend to shorter overall survival.
Using immunohistochemistry, Ougolkov et al[96] could show nuclear overexpression of EZH2 in pancreatic cancer cell lines and in 68% (71/104) of pancreatic adenocarcinomas and that its nuclear accumulation was more prevalent in poorly differentiated pancreatic adenocarcinomas (91%, 31/34). Using RNA interference, genetic depletion of EZH2 sensitized pancreatic cancer cells to chemotherapy and caused re-expression of p27 [cyclin-dependent kinase inhibitor 1B (p27, Kip1)] and decreased proliferation. Similar results were obtained by Toll et al[97] who demonstrated an inverse relationship between expression of EZH2 and E-cadherin in 54 pancreatic adenocarcinomas, and significantly longer survival in gemcitabine-treated patients with low expression of EZH2. Interestingly, inactivation of RUNX3 (runt-related transcription factor 3), a component of TGF-β signalling, is mediated by at least 2 epigenetic mechanisms, both EZH2[98] and DNA hypermethylation[99], highlighting their cooperation to convey a malignant phenotype in pancreatic cancer. Recently, miR-218 was demonstrated to be negatively regulated by EZH2 in pancreatic ductal adenocarcinoma[100]. MiR-218 was significantly reduced in primary tumor samples compared with normal adjacent tissue, and its silencing was mediated by EZH2 which binds to the miR-218 promoter, promotes formation of heterochromatin, and recruits DNMT-1, -3A and -3B. MiR-218 expression reduces proliferation in vitro and tumor formation as well as metastasis in nude mice[100]. The inverse regulatory relationship has also been recently observed between EZH2 and pre-miR101. Overexpression of pre-miR-101 reduced the binding of EZH2 to the promoter of the E-cadherin gene (CDH1) and increased the levels of E-cadherin[101].
Epigenetics in pancreatic cancer stem cells: An interesting and possibly therapeutically relevant aspect is the role of EZH2 in maintenance of stemness characteristics in cancer, particular its role in maintaining the self-renewal capabilities of cancer stem cells (CSC)[102-108]. This subpopulation of cancer cells has been characterized in pancreatic cancer by surface markers CD44, CD24, CD133, ESA (EpCAM, epithelial cell adhesion molecule)[109,110] and is thought to represent the population of cancer cells responsible for tumor maintenance, tumorigenicity, metastasis, and resistance to conventional chemotherapeutic drugs, as well as recurrence[109,111]. Similar to studies with CSC derived from hepatocellular carcinoma and acute myeloid leukemia[104,112], recent studies have demonstrated the therapeutic potential of epidrugs to directly target this tumorigenic subpopulation of pancreatic cancer cells: Avan et al[102] used an inhibitor of the EZH2 methyltransferase (DZNep, deazaneplanocin-A) and showed that treatment with DZNep reduced spheroid formation of pancreatic cancer cells and decreased the CD133+ subpopulation. Furthermore, the combination of DZNep and gemcitabine was shown to be highly synergistic and was accompanied by a reduced percentage of G2/M cells, reduced migration, increased E-cadherin expression and increased apoptosis[102]. The level of EZH2 has furthermore been suggested as an assay to effectively measure changes in the CSC subpopulation: using pancreatic and breast cancer cell lines, knockdown of EZH2 by RNA interference decreased the CSC subpopulation, confirming its role in CSC maintenance, and genes affected by EZH2 knockdown were inversely correlated with their expression in enriched CSC subpopulations[108]. The Hedgehog pathway has also been implicated in the maintenance of CSCs in various models[9,113]; interestingly a combination of Hedgehog inhibition (SANT-1) and SAHA (a pan-HDAC inhibitor; suberanilohydroxamic acid, Vorinostat) synergistically suppressed proliferation and colony formation in gemcitabine-resistant pancreatic adenocarcinoma cell lines by increased Bax expression, activation of caspase-3/7, increased p21 and p27 and reduced cyclin D1 expression. This study suggests that combined inhibition of stem cell-associated pathways (Hedgehog) and epigenetic drugs could be efficient in targeting the CSC subpopulation in pancreatic cancer[114]. A study by Nalls et al[115] could demonstrate that demethylating agents (5-aza-dC, 5-aza-2’-deoxycytidine) and the HDAC inhibitor SAHA restored the expression levels of miR-34a, which is reduced in pancreatic CSCs. These inhibitors caused a reduction in the EMT-related ZEB1, Snail, and Slug transcription factors, increased epithelial marker expression (E-cadherin) and, most importantly, reduced the number of viable pancreatic CSC, accompanied by reduced migration, colony formation and invasion of these cells.
Based on the above-mentioned functions and properties of CSC which is critical for tumor initiation, metastasis, progression and therapeutic resistance, these findings are of central importance and warrant further investigation to hopefully develop (epigenetics-based) therapeutic regimens specifically targeting this tumorigenic subpopulation in pancreatic cancer.
miRNA-based epigenetics in pancreatic cancer: Some reporters[116-119] and Park et al[120] have reviewed the available publications on differential miRNA expression in pancreatic cancer vs normal tissue culminating in a list of 64, partly overlapping individual miRNAs which were found to be deregulated in pancreatic cancer. Of these miRNAs, overexpression of miR-21, -155, -196a-2, -203, -210 and -222 was furthermore associated with poor outcome[120].
Table 4 provides an update (based on Park et al[120] 2011) of the currently available literature on the specific role of individual miRNAs in pancreatic cancer. All of these studies investigated the cellular/molecular mechanisms of the oncogenic or tumor-suppressive action of miRNAs, mainly by forced overexpression or knockdown of the respective miRNAs.
An example of how epigenetic mechanisms are employed to regulate the expression of tumor-suppressive miRNAs is shown in the study of Zhang et al[121]. From 12 miRNAs differentially expressed in pancreatic cancers vs adjacent normal tissue, miR-132 was downregulated in 16/20 pancreatic carcinomas accompanied by methylation of its promoter, as shown both in cell lines and tumor tissue. Sp-1 expression was correlated with miR-132 expression, and its binding affinity to the miR-132 promoter was significantly lower in pancreatic tumors relative to non-tumor samples.
As recently discussed[120,122], epigenetic features and especially miRNAs could also serve as biomarkers to allow specific and sensitive diagnosis of pancreatic cancer - an important approach as most patients with this disease remain without symptoms until the lesion has progressed to an advanced or metastatic stage. In this context, the use of miR-155 which showed upregulation in most IPMNs (83% of cases) has been analyzed in pancreatic juice by Habbe et al[123]. The authors confirmed upregulation of the miR-155 transcript in 60% (6/10) of IPMN-associated pancreatic juice samples but in none of the 5 control cases. Wang et al[124] profiled 4 miRNAs (miR-21, -210, -155, and -196a) in heparin-treated blood samples and found a sensitivity of 64% and a specificity of 89% to distinguish pancreatic cancer patients from healthy controls using this panel of miRNAs, thus proving the feasibility of plasma-based miRNA profiling as a potential biomarker for pancreatic cancer. Furthermore, Kawaguchi et al[125] investigated the utility of plasma miR-221 as a biomarker for cancer detection and monitoring tumor dynamics in 47 consecutive pancreatic cancer patients: similar to cancer tissue, plasma miR-221 levels were significantly higher in pancreatic cancer patients and correlated with distant metastasis and non-resectable status. Also, miR-21 serum levels were shown to be associated with overall survival of pancreatic cancer patients, and, in combination with 6 other miRNAs, allowed for correct classification of clinically suspected pancreatic cancer with a rate of 84%[126]. Similar results were obtained for miR-18a in plasma samples of patients with pancreatic cancer: miR-18a levels were significantly higher in 36 cancer patients compared with 30 healthy volunteers[127]. Kong et al[128] investigated the utility of several miRNAs as serum markers: while miR-21 distinguished pancreatic ductal adenocarcinoma patients from chronic pancreatitis and controls, miR-196a could distinguish resectable (stages I and II) and unresectable pancreatic ductal adenocarcinoma (III and IV) as well as predict median survival time of pancreatic ductal adenocarcinoma patients (6.1 mo vs 12.0 mo for high vs low level miR-196a). Recently, miR-21 from pancreatic cyst fluid was investigated as a potential biomarker and could differentiate between benign, premalignant and malignant pancreatic cyst neoplasms[129].
The classic cancer progression model from PanIN to invasive carcinoma highlights genetic alterations in several oncogenes and tumor suppressor genes[130]. Hanahan et al[35,36] characterized additional distinct features of malignant tumor cells in their outstanding reviews on hallmarks of cancer that have also been identified in pancreatic cancer: sustaining proliferative signaling (e.g., activating mutations of K-ras[131]), evading growth suppressors (e.g., deletions or mutations of CDKN2A/p16Ink4A[132]), activating invasion and metastasis (e.g., expression of CXCL12/CXCR4 [chemokine (C-X-C motif) ligand 12/(CXC receptor 4)[133]], enabling replicative immortality (e.g., telomerase activation via loss of ATRX in pancreatic neuroendocrine tumors[134]), inducing angiogenesis [e.g., increase in serum vascular endothelial growth factor (VEGF)[135]] and resisting cell death (e.g., overexpression of anti-apoptotic Bcl-2[136]. Many of these alterations have been explored as targets for novel therapies (e.g., anti-angiogenesis using the anti-VEGF antibody bevacizumab or anti-epidermal growth factor receptor directed therapies using erlotinib or cetuximab) achieved only marginal survival benefits in pancreatic cancer patients compared with standard therapy[137-139]. As outlined above, recent data also suggest strong roles for non-genetic events in pancreatic carcinogenesis and resistance to current therapies[8], e.g., by modulating ABC drug transporters or interfering with cell death pathways (see Tables 2-4 for details).
Consequently, these regulatory mechanisms could represent interesting and potent novel targets for therapy to overcome resistance and to improve treatment outcome further[140,141]. Inhibitors of DNMT are nucleoside analogues of cytidine and currently azacytidine and decitabine are available for clinical use (Table 6), although the number of current trials is very limited. Zebularine is in preclinical development[142] with promising experimental data in pancreatic cancer[143].
| Compound | Combination | Phase | Endpoint | ClincialTrials.gov | Treatment |
| DNMT inhibitors | |||||
| Azacitidine | + Gemcitabine | I | MTD | NCT01167816 | |
| Azacitidine | II | PFS | NCT01845805 | 2 | |
| Decitabine | Various stages of development for solid tumors | 3 | |||
| Zebularine | Preclinical | ||||
| HDAC inhibitors | |||||
| Vorinostat | + Marizomib | I | MTD | NCT00667082[234] | |
| Vorinostat | + Radiation | I/II | MTD, PFS | NCT00948688 | |
| + 5-FU | |||||
| Vorinostat | + Radiation | I | MTD | NCT00983268 | |
| + Capecitabine | |||||
| Vorinostat | + Radiation | I/II | MTD | NCT00831493 | |
| Belinostat | Various stages of development for solid tumors | ||||
| Entinostat | I | MTD | NCT00020579[235] | ||
| Entinostat | 13-cis retinoic acid | I | MTD | ||
| Panobinostat | + Bortezomib | II | PFS | NCT01056601[236] | 1 |
| HAT inhibitors | |||||
| Curcumin | II | Survival | NCT00094445[147] | 1 | |
| Curcumin | + Gemcitabine | II | TTP | NCT00192842 | 1 |
| Curcumin | + Gemcitabine | III | NCT00486460 | 1 | |
| + Celecoxib | |||||
| Curcumin | I | MTD | -[237] | ||
| Curcumin | + Gemcitabine | I | MTD | -[238] | |
| Curcumin | + Gemcitabine | I/II | MTD | -[239] | |
Inhibitors of protein and histone deacetylases have been established as a novel approach to target hematologic and solid tumors[144]. Several phase I studies using the first-in-class molecule vorinostat (SAHA) are currently ongoing, especially in combination with cytotoxic agents or radiotherapy. Other agents like belinostat (PXD-101), entinostat (MS-275) or panobinostat (LBH-589) are at various stages of early clinical development, too, with progression-free survival or maximum tolerated dose as study endpoints. As described above, in addition to deacetylases, HATs can also regulate gene transcription. Here, curcumin (derived from the South Asian plant turmeric) has been demonstrated to effectively inhibit the activity of the HAT p300/CBP in cancer cells[145,146]. Although its pharmacokinetic properties are unsatisfying so far, it demonstrated early signs of clinical efficacy in pancreatic cancer patients in a phase II setting[147].
Other epigenetic modifiers besides DNMT, HAT or HDAC have been identified and the first lead compounds are currently being extensively studied preclinically or are in early clinical phases. However, clinical data for pancreatic cancer is not available[148].
While miRNAs are considered useful tools for diagnosis, prognosis and possibly patient stratification[149], miRNA-based therapeutics are currently not available. Although preclinical data suggests that antagomiRs or miRNA replacement therapy is promising for pancreatic cancer models, clinical use is hampered by unresolved drug delivery and the fact that one miRNA also has several target mRNAs, thus possibly being too unspecific[150,151].
Overall, as most of the agents highlighted above are currently in early phases of clinical development, no clear data on efficacy of epigenetic agents in pancreatic cancer are available, but promising preclinical[152] and early clinical data warrant further development.
Due to the poor prognosis of pancreatic cancer patients, understanding the molecular events driving this devastating tumor disease is central for development of alternative and more effective treatment strategies and for the determination of reliable diagnostic markers. Recent research on epigenetic mechanisms has greatly enriched our knowledge about the regulatory traits involved in initiation, progression and metastasis of pancreatic cancer. As reviewed in this article, DNA-, histone- and miRNA-based epigenetic events have been demonstrated to play a role in pancreatic cancer and could serve as future therapeutic targets aiming at reversing the epigenetic deregulation of the cellular machinery. Initial clinical trials at stages I-III using inhibitors of DNMTs, HDACs and HATs are currently under way and open the door to development of novel and hopefully more effective ‘epidrugs’ for patients with pancreatic cancer.
Ocker M is an employee of Bayer Pharma AG.
P- Reviewers: Barreto S, Caswell PT, Gu DS, Nakano H, Yang F S- Editor: Wen LL L- Editor: Cant MR E- Editor: Zhang DN
| 1. | Ma J, Siegel R, Jemal A. Pancreatic cancer death rates by race among US men and women, 1970-2009. J Natl Cancer Inst. 2013;105:1694-1700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 115] [Article Influence: 10.5] [Reference Citation Analysis (1)] |
| 2. | Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62:317-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 3. | Li D, Abbruzzese JL. New strategies in pancreatic cancer: emerging epidemiologic and therapeutic concepts. Clin Cancer Res. 2010;16:4313-4318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Herreros-Villanueva M, Hijona E, Cosme A, Bujanda L. Adjuvant and neoadjuvant treatment in pancreatic cancer. World J Gastroenterol. 2012;18:1565-1572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 41] [Cited by in F6Publishing: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Kim SC, Kim YH, Park KM, Lee YJ. Pancreatic cancer surgery: the state of the art. Curr Drug Targets. 2012;13:764-771. [PubMed] [Cited in This Article: ] |
| 6. | Jarboe J, Saif MW. First line therapy for metastatic pancreatic cancer. JOP. 2013;14:340-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 7. | Carpelan-Holmström M, Nordling S, Pukkala E, Sankila R, Lüttges J, Klöppel G, Haglund C. Does anyone survive pancreatic ductal adenocarcinoma A nationwide study re-evaluating the data of the Finnish Cancer Registry. Gut. 2005;54:385-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 255] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 8. | McCleary-Wheeler AL, Lomberk GA, Weiss FU, Schneider G, Fabbri M, Poshusta TL, Dusetti NJ, Baumgart S, Iovanna JL, Ellenrieder V. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett. 2013;328:212-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Kiesslich T, Berr F, Alinger B, Kemmerling R, Pichler M, Ocker M, Neureiter D. Current status of therapeutic targeting of developmental signalling pathways in oncology. Curr Pharm Biotechnol. 2012;13:2184-2220. [PubMed] [Cited in This Article: ] |
| 10. | Quint K, Stintzing S, Alinger B, Hauser-Kronberger C, Dietze O, Gahr S, Hahn EG, Ocker M, Neureiter D. The expression pattern of PDX-1, SHH, Patched and Gli-1 is associated with pathological and clinical features in human pancreatic cancer. Pancreatology. 2009;9:116-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 12. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25175] [Article Influence: 1936.5] [Reference Citation Analysis (3)] |
| 13. | American Society of Clinical Oncology (ASCO): Cancer Net. Accessed Nov 27. 2013; Available from: http://www.cancer.net/cancer-types/pancreatic-cancer/statistics.. [Cited in This Article: ] |
| 14. | Schnelldorfer T, Ware AL, Sarr MG, Smyrk TC, Zhang L, Qin R, Gullerud RE, Donohue JH, Nagorney DM, Farnell MB. Long-term survival after pancreatoduodenectomy for pancreatic adenocarcinoma: is cure possible. Ann Surg. 2008;247:456-462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 339] [Cited by in F6Publishing: 352] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 15. | Lüttges J, Schemm S, Vogel I, Hedderich J, Kremer B, Klöppel G. The grade of pancreatic ductal carcinoma is an independent prognostic factor and is superior to the immunohistochemical assessment of proliferation. J Pathol. 2000;191:154-161. [PubMed] [Cited in This Article: ] |
| 16. | Klöppel G, Lingenthal G, von Bülow M, Kern HF. Histological and fine structural features of pancreatic ductal adenocarcinomas in relation to growth and prognosis: studies in xenografted tumours and clinico-histopathological correlation in a series of 75 cases. Histopathology. 1985;9:841-856. [PubMed] [Cited in This Article: ] |
| 17. | Giovinazzo F, Turri G, Zanini S, Butturini G, Scarpa A, Bassi C. Clinical implications of biological markers in Pancreatic Ductal Adenocarcinoma. Surg Oncol. 2012;21:e171-e182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Adsay NV, Basturk O, Bonnett M, Kilinc N, Andea AA, Feng J, Che M, Aulicino MR, Levi E, Cheng JD. A proposal for a new and more practical grading scheme for pancreatic ductal adenocarcinoma. Am J Surg Pathol. 2005;29:724-733. [PubMed] [Cited in This Article: ] |
| 19. | Karamitopoulou E. Role of Epithelial-Mesenchymal Transition in Pancreatic Ductal Adenocarcinoma: Is Tumor Budding the Missing Link. Front Oncol. 2013;3:221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 20. | Handra-Luca A, Hong SM, Walter K, Wolfgang C, Hruban R, Goggins M. Tumour epithelial vimentin expression and outcome of pancreatic ductal adenocarcinomas. Br J Cancer. 2011;104:1296-1302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 21. | Wang WY, Hsu CC, Wang TY, Li CR, Hou YC, Chu JM, Lee CT, Liu MS, Su JJ, Jian KY. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology. 2013;145:1110-1120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Hruban RH, Adsay NV. Molecular classification of neoplasms of the pancreas. Hum Pathol. 2009;40:612-623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, Sohn TA, Kadkol SS, Yeo CJ, Choti M, Zahurak M. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol. 2000;156:1641-1651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 161] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 24. | Zamboni G, Hirabayashi K, Castelli P, Lennon AM. Precancerous lesions of the pancreas. Best Pract Res Clin Gastroenterol. 2013;27:299-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin. Cancer Cell. 2012;22:9-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 794] [Cited by in F6Publishing: 749] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 26. | Breitkreutz D, Hlatky L, Rietman E, Tuszynski JA. Molecular signaling network complexity is correlated with cancer patient survivability. Proc Natl Acad Sci USA. 2012;109:9209-9212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 27. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2858] [Cited by in F6Publishing: 2881] [Article Influence: 180.1] [Reference Citation Analysis (0)] |
| 28. | Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 628] [Cited by in F6Publishing: 569] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 29. | Ueki T, Toyota M, Skinner H, Walter KM, Yeo CJ, Issa JP, Hruban RH, Goggins M. Identification and characterization of differentially methylated CpG islands in pancreatic carcinoma. Cancer Res. 2001;61:8540-8546. [PubMed] [Cited in This Article: ] |
| 30. | Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735-3742. [PubMed] [Cited in This Article: ] |
| 31. | Hong SM, Park JY, Hruban RH, Goggins M. Molecular signatures of pancreatic cancer. Arch Pathol Lab Med. 2011;135:716-727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 15] [Reference Citation Analysis (0)] |
| 32. | Yamamoto H, Itoh F, Nakamura H, Fukushima H, Sasaki S, Perucho M, Imai K. Genetic and clinical features of human pancreatic ductal adenocarcinomas with widespread microsatellite instability. Cancer Res. 2001;61:3139-3144. [PubMed] [Cited in This Article: ] |
| 33. | Nakata B, Wang YQ, Yashiro M, Nishioka N, Tanaka H, Ohira M, Ishikawa T, Nishino H, Hirakawa K. Prognostic value of microsatellite instability in resectable pancreatic cancer. Clin Cancer Res. 2002;8:2536-2540. [PubMed] [Cited in This Article: ] |
| 34. | Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835-1839. [PubMed] [Cited in This Article: ] |
| 35. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [PubMed] [Cited in This Article: ] |
| 36. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42956] [Article Influence: 3304.3] [Reference Citation Analysis (4)] |
| 37. | Gnoni A, Licchetta A, Scarpa A, Azzariti A, Brunetti AE, Simone G, Nardulli P, Santini D, Aieta M, Delcuratolo S. Carcinogenesis of pancreatic adenocarcinoma: precursor lesions. Int J Mol Sci. 2013;14:19731-19762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 38. | Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163-170. [PubMed] [Cited in This Article: ] |
| 39. | Grzenda A, Ordog T, Urrutia R. Polycomb and the emerging epigenetics of pancreatic cancer. J Gastrointest Cancer. 2011;42:100-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 40. | Lomberk GA. Epigenetic silencing of tumor suppressor genes in pancreatic cancer. J Gastrointest Cancer. 2011;42:93-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Antequera F, Bird A. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol. 1999;9:R661-R667. [PubMed] [Cited in This Article: ] |
| 42. | Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 269] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 43. | Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89-92. [PubMed] [Cited in This Article: ] |
| 44. | Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1515] [Cited by in F6Publishing: 1449] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 45. | Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2672] [Cited by in F6Publishing: 2493] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 46. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6156] [Cited by in F6Publishing: 5912] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 47. | Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285-294. [PubMed] [Cited in This Article: ] |
| 48. | Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037-1043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 251] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 49. | Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074-1080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7019] [Cited by in F6Publishing: 6658] [Article Influence: 289.5] [Reference Citation Analysis (0)] |
| 50. | Bojang P, Ramos KS. The promise and failures of epigenetic therapies for cancer treatment. Cancer Treat Rev. 2014;40:153-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 51. | Koutsounas I, Giaginis C, Patsouris E, Theocharis S. Current evidence for histone deacetylase inhibitors in pancreatic cancer. World J Gastroenterol. 2013;19:813-828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 33] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 52. | Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257-267. [PubMed] [Cited in This Article: ] |
| 53. | Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470-479. [PubMed] [Cited in This Article: ] |
| 54. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14460] [Cited by in F6Publishing: 15327] [Article Influence: 1021.8] [Reference Citation Analysis (1)] |
| 55. | Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83-92. [PubMed] [Cited in This Article: ] |
| 56. | Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817-1821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1137] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 57. | Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959-5974. [PubMed] [Cited in This Article: ] |
| 58. | Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-D144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3282] [Cited by in F6Publishing: 3401] [Article Influence: 188.9] [Reference Citation Analysis (0)] |
| 59. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5833] [Cited by in F6Publishing: 6223] [Article Influence: 388.9] [Reference Citation Analysis (0)] |
| 60. | Xi JJ. MicroRNAs in Cancer. Cancer Treat Res. 2013;158:119-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Melo SA, Kalluri R. Molecular pathways: microRNAs as cancer therapeutics. Clin Cancer Res. 2012;18:4234-4239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 62. | Perera RJ, Ray A. Epigenetic regulation of miRNA genes and their role in human melanomas. Epigenomics. 2012;4:81-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 63. | Agirre X, Martínez-Climent JÁ, Odero MD, Prósper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. 2012;26:395-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 64. | Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM, Ausseil F, Vispé S, Arimondo PB. DNA methylation inhibitors in cancer: recent and future approaches. Biochimie. 2012;94:2280-2296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 169] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 65. | Peedicayil J. The role of DNA methylation in the pathogenesis and treatment of cancer. Curr Clin Pharmacol. 2012;7:333-340. [PubMed] [Cited in This Article: ] |
| 66. | Mikeska T, Bock C, Do H, Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn. 2012;12:473-487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 67. | Orr JA, Hamilton PW. Histone acetylation and chromatin pattern in cancer. A review. Anal Quant Cytol Histol. 2007;29:17-31. [PubMed] [Cited in This Article: ] |
| 68. | Shukla V, Vaissière T, Herceg Z. Histone acetylation and chromatin signature in stem cell identity and cancer. Mutat Res. 2008;637:1-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 69. | Mai A, Massa S, Rotili D, Cerbara I, Valente S, Pezzi R, Simeoni S, Ragno R. Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev. 2005;25:261-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 248] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 70. | Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkühler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 2007;17:195-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 362] [Cited by in F6Publishing: 398] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 71. | Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 72. | Decarlo D, Hadden MK. Oncoepigenomics: making histone lysine methylation count. Eur J Med Chem. 2012;56:179-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 73. | Tian X, Zhang S, Liu HM, Zhang YB, Blair CA, Mercola D, Sassone-Corsi P, Zi X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013;13:558-579. [PubMed] [Cited in This Article: ] |
| 74. | Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 463] [Cited by in F6Publishing: 550] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 75. | Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125-136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 186] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 76. | Hoshino I, Matsubara H. MicroRNAs in cancer diagnosis and therapy: from bench to bedside. Surg Today. 2013;43:467-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 77. | Zhu Y, Zhang JJ, Zhu R, Zhu Y, Liang WB, Gao WT, Yu JB, Xu ZK, Miao Y. The increase in the expression and hypomethylation of MUC4 gene with the progression of pancreatic ductal adenocarcinoma. Med Oncol. 2011;28 Suppl 1:S175-S184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 78. | Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158-4166. [PubMed] [Cited in This Article: ] |
| 79. | Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, Savoy DN, Molina JR, Fonseca R, Smyrk TC, Chari ST, Urrutia R, Billadeau DD. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 175] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 80. | He S, Wang F, Yang L, Guo C, Wan R, Ke A, Xu L, Hu G, Xu X, Shen J. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS One. 2011;6:e27684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 81. | Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115-2125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 203] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 82. | Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68:2708-2716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 83. | Yamada N, Hamada T, Goto M, Tsutsumida H, Higashi M, Nomoto M, Yonezawa S. MUC2 expression is regulated by histone H3 modification and DNA methylation in pancreatic cancer. Int J Cancer. 2006;119:1850-1857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 84. | Yamada N, Nishida Y, Yokoyama S, Tsutsumida H, Houjou I, Kitamoto S, Goto M, Higashi M, Yonezawa S. Expression of MUC5AC, an early marker of pancreatobiliary cancer, is regulated by DNA methylation in the distal promoter region in cancer cells. J Hepatobiliary Pancreat Sci. 2010;17:844-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 85. | Fritsche P, Seidler B, Schüler S, Schnieke A, Göttlicher M, Schmid RM, Saur D, Schneider G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 2009;58:1399-1409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 86. | Ouaïssi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirrò N, Seitz JF. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318-2328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 87. | Schneider G, Krämer OH, Fritsche P, Schüler S, Schmid RM, Saur D. Targeting histone deacetylases in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2010;14:1255-1263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 88. | Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Büchler M, Evert M, Lerch MM. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439-448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 89. | Schneider G, Krämer OH, Saur D. A ZEB1-HDAC pathway enters the epithelial to mesenchymal transition world in pancreatic cancer. Gut. 2012;61:329-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 90. | von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, von Werder A, Schmidt A, Mages J, Pagel P. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361-71, 371.e1-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 276] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 91. | Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application. Crit Rev Oncol Hematol. 2012;83:184-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 92. | Martínez-Romero C, Rooman I, Skoudy A, Guerra C, Molero X, González A, Iglesias M, Lobato T, Bosch A, Barbacid M. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. J Pathol. 2009;219:205-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 93. | Song W, Tao K, Li H, Jin C, Song Z, Li J, Shi H, Li X, Dang Z, Dou K. Bmi-1 is related to proliferation, survival and poor prognosis in pancreatic cancer. Cancer Sci. 2010;101:1754-1760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 94. | Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487-1495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1262] [Cited by in F6Publishing: 1339] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 95. | Karamitopoulou E, Pallante P, Zlobec I, Tornillo L, Carafa V, Schaffner T, Borner M, Diamantis I, Esposito F, Brunner T. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer. 2010;46:1438-1444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 96. | Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 2008;14:6790-6796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 97. | Toll AD, Dasgupta A, Potoczek M, Yeo CJ, Kleer CG, Brody JR, Witkiewicz AK. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41:1205-1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 98. | Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283:17324-17332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 99. | Nomoto S, Kinoshita T, Mori T, Kato K, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Adverse prognosis of epigenetic inactivation in RUNX3 gene at 1p36 in human pancreatic cancer. Br J Cancer. 2008;98:1690-1695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 100. | Li CH, To KF, Tong JH, Xiao Z, Xia T, Lai PB, Chow SC, Zhu YX, Chan SL, Marquez VE. Enhancer of zeste homolog 2 silences microRNA-218 in human pancreatic ductal adenocarcinoma cells by inducing formation of heterochromatin. Gastroenterology. 2013;144:1086-1097.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 101. | Qazi AM, Gruzdyn O, Semaan A, Seward S, Chamala S, Dhulipala V, Sethi S, Ali-Fehmi R, Philip PA, Bouwman DL. Restoration of E-cadherin expression in pancreatic ductal adenocarcinoma treated with microRNA-101. Surgery. 2012;152:704-11; discussion 711-3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 102. | Avan A, Crea F, Paolicchi E, Funel N, Galvani E, Marquez VE, Honeywell RJ, Danesi R, Peters GJ, Giovannetti E. Molecular mechanisms involved in the synergistic interaction of the EZH2 inhibitor 3-deazaneplanocin A with gemcitabine in pancreatic cancer cells. Mol Cancer Ther. 2012;11:1735-1746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 103. | Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 324] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 104. | Chiba T, Suzuki E, Negishi M, Saraya A, Miyagi S, Konuma T, Tanaka S, Tada M, Kanai F, Imazeki F. 3-Deazaneplanocin A is a promising therapeutic agent for the eradication of tumor-initiating hepatocellular carcinoma cells. Int J Cancer. 2012;130:2557-2567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 105. | Crea F, Hurt EM, Mathews LA, Cabarcas SM, Sun L, Marquez VE, Danesi R, Farrar WL. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol Cancer. 2011;10:40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 106. | Rizzo S, Hersey JM, Mellor P, Dai W, Santos-Silva A, Liber D, Luk L, Titley I, Carden CP, Box G. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol Cancer Ther. 2011;10:325-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 107. | Stefansson OA, Esteller M. EZH2-mediated epigenetic repression of DNA repair in promoting breast tumor initiating cells. Breast Cancer Res. 2011;13:309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 108. | van Vlerken LE, Kiefer CM, Morehouse C, Li Y, Groves C, Wilson SD, Yao Y, Hollingsworth RE, Hurt EM. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med. 2013;2:43-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 109. | Dorado J, Lonardo E, Miranda-Lorenzo I, Heeschen C. Pancreatic cancer stem cells: new insights and perspectives. J Gastroenterol. 2011;46:966-973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 110. | Li C, Lee CJ, Simeone DM. Identification of human pancreatic cancer stem cells. Methods Mol Biol. 2009;568:161-173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 179] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 111. | Alison MR, Lim SM, Nicholson LJ. Cancer stem cells: problems for therapy. J Pathol. 2011;223:147-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 219] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 112. | Zhou J, Bi C, Cheong LL, Mahara S, Liu SC, Tay KG, Koh TL, Yu Q, Chng WJ. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood. 2011;118:2830-2839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 174] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 113. | Merchant AA, Matsui W. Targeting Hedgehog--a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130-3140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 379] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 114. | Chun SG, Zhou W, Yee NS. Combined targeting of histone deacetylases and hedgehog signaling enhances cytoxicity in pancreatic cancer. Cancer Biol Ther. 2009;8:1328-1339. [PubMed] [Cited in This Article: ] |
| 115. | Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 207] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 116. | Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 216] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 117. | Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901-1908. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 873] [Cited by in F6Publishing: 952] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 118. | Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 520] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 119. | Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046-1054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 632] [Cited by in F6Publishing: 679] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 120. | Park JY, Helm J, Coppola D, Kim D, Malafa M, Kim SJ. MicroRNAs in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2011;17:817-827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 27] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 121. | Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183-1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 122. | Fukushige S, Horii A. Road to early detection of pancreatic cancer: Attempts to utilize epigenetic biomarkers. Cancer Lett. 2014;342:231-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 123. | Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340-346. [PubMed] [Cited in This Article: ] |
| 124. | Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2:807-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 418] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 125. | Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 126. | Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 294] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 127. | Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733-1740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 128. | Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 129. | Farrell JJ, Toste P, Wu N, Li L, Wong J, Malkhassian D, Tran LM, Wu X, Li X, Dawson D. Endoscopically acquired pancreatic cyst fluid microRNA 21 and 221 are associated with invasive cancer. Am J Gastroenterol. 2013;108:1352-1359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 130. | Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002-2006. [PubMed] [Cited in This Article: ] |
| 131. | Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545-554. [PubMed] [Cited in This Article: ] |
| 132. | Rachakonda PS, Bauer AS, Xie H, Campa D, Rizzato C, Canzian F, Beghelli S, Greenhalf W, Costello E, Schanne M. Somatic mutations in exocrine pancreatic tumors: association with patient survival. PLoS One. 2013;8:e60870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 133. | Zhong W, Chen W, Zhang D, Sun J, Li Y, Zhang J, Gao Y, Zhou W, Li S. CXCL12/CXCR4 axis plays pivotal roles in the organ-specific metastasis of pancreatic adenocarcinoma: A clinical study. Exp Ther Med. 2012;4:363-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 134. | Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 844] [Cited by in F6Publishing: 782] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 135. | Sivaprasad S, Govardhan B, Harithakrishna R, Venkat Rao G, Pradeep R, Kunal B, Ramakrishna N, Anuradha S, Reddy DN. Association of vascular endothelial growth factor (VEGF) gene polymorphism and increased serum VEGF concentration with pancreatic adenocarcinoma. Pancreatology. 2013;13:267-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 136. | Sun CY, Wang BL, Hu CQ, Peng RY, Gao YB, Gu QY, Wang DW. Expression of the bcl-2 gene and its significance in human pancreatic carcinoma. Hepatobiliary Pancreat Dis Int. 2002;1:306-308. [PubMed] [Cited in This Article: ] |
| 137. | Philip PA, Benedetti J, Corless CL, Wong R, O’Reilly EM, Flynn PJ, Rowland KM, Atkins JN, Mirtsching BC, Rivkin SE. Phase III study comparing gemcitabine plus cetuximab versus gemcitabine in patients with advanced pancreatic adenocarcinoma: Southwest Oncology Group-directed intergroup trial S0205. J Clin Oncol. 2010;28:3605-3610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 478] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 138. | Kindler HL, Niedzwiecki D, Hollis D, Sutherland S, Schrag D, Hurwitz H, Innocenti F, Mulcahy MF, O’Reilly E, Wozniak TF. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol. 2010;28:3617-3622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 645] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 139. | Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960-1966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2755] [Cited by in F6Publishing: 2667] [Article Influence: 156.9] [Reference Citation Analysis (0)] |
| 140. | Stintzing S, Kemmerling R, Kiesslich T, Alinger B, Ocker M, Neureiter D. Myelodysplastic syndrome and histone deacetylase inhibitors: “to be or not to be acetylated”. J Biomed Biotechnol. 2011;2011:214143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 141. | Schneider-Stock R, Ocker M. Epigenetic therapy in cancer: molecular background and clinical development of histone deacetylase and DNA methyltransferase inhibitors. IDrugs. 2007;10:557-561. [PubMed] [Cited in This Article: ] |
| 142. | Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33:2989-2996. [PubMed] [Cited in This Article: ] |
| 143. | Neureiter D, Zopf S, Leu T, Dietze O, Hauser-Kronberger C, Hahn EG, Herold C, Ocker M. Apoptosis, proliferation and differentiation patterns are influenced by Zebularine and SAHA in pancreatic cancer models. Scand J Gastroenterol. 2007;42:103-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 144. | Ocker M. Deacetylase inhibitors - focus on non-histone targets and effects. World J Biol Chem. 2010;1:55-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 47] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 145. | Fu S, Kurzrock R. Development of curcumin as an epigenetic agent. Cancer. 2010;116:4670-4676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 146. | Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169-174. [PubMed] [Cited in This Article: ] |
| 147. | Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, Ng CS, Badmaev V, Kurzrock R. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491-4499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 880] [Cited by in F6Publishing: 932] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 148. | Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765-780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 149. | Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, Barth RJ, Suriawinata AA, Tsongalis GJ. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013;206:217-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 150. | Khan S, Ansarullah D, Jaggi M, Chauhan SC. Targeting microRNAs in pancreatic cancer: microplayers in the big game. Cancer Res. 2013;73:6541-6547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 151. | Pai P, Rachagani S, Are C, Batra SK. Prospects of miRNA-based therapy for pancreatic cancer. Curr Drug Targets. 2013;14:1101-1109. [PubMed] [Cited in This Article: ] |
| 152. | Gahr S, Ocker M, Ganslmayer M, Zopf S, Okamoto K, Hartl A, Leitner S, Hahn EG, Herold C. The combination of the histone-deacetylase inhibitor trichostatin A and gemcitabine induces inhibition of proliferation and increased apoptosis in pancreatic carcinoma cells. Int J Oncol. 2007;31:567-576. [PubMed] [Cited in This Article: ] |
| 153. | Adsay NV, Pierson C, Sarkar F, Abrams J, Weaver D, Conlon KC, Brennan MF, Klimstra DS. Colloid (mucinous noncystic) carcinoma of the pancreas. Am J Surg Pathol. 2001;25:26-42. [PubMed] [Cited in This Article: ] |
| 154. | Brody JR, Costantino CL, Potoczek M, Cozzitorto J, McCue P, Yeo CJ, Hruban RH, Witkiewicz AK. Adenosquamous carcinoma of the pancreas harbors KRAS2, DPC4 and TP53 molecular alterations similar to pancreatic ductal adenocarcinoma. Mod Pathol. 2009;22:651-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 155. | Hoorens A, Prenzel K, Lemoine NR, Klöppel G. Undifferentiated carcinoma of the pancreas: analysis of intermediate filament profile and Ki-ras mutations provides evidence of a ductal origin. J Pathol. 1998;185:53-60. [PubMed] [Cited in This Article: ] |
| 156. | Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 160] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 157. | Hong SM, Kelly D, Griffith M, Omura N, Li A, Li CP, Hruban RH, Goggins M. Multiple genes are hypermethylated in intraductal papillary mucinous neoplasms of the pancreas. Mod Pathol. 2008;21:1499-1507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 158. | Kim SG, Wu TT, Lee JH, Yun YK, Issa JP, Hamilton SR, Rashid A. Comparison of epigenetic and genetic alterations in mucinous cystic neoplasm and serous microcystic adenoma of pancreas. Mod Pathol. 2003;16:1086-1094. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 159. | Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21:238-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 160. | Sato N, Fukushima N, Matsubayashi H, Iacobuzio-Donahue CA, Yeo CJ, Goggins M. Aberrant methylation of Reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma. Cancer. 2006;107:251-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 161. | Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2002;123:365-372. [PubMed] [Cited in This Article: ] |
| 162. | Ruggeri BA, Huang L, Wood M, Cheng JQ, Testa JR. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81-86. [PubMed] [Cited in This Article: ] |
| 163. | Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636-3641. [PubMed] [Cited in This Article: ] |
| 164. | Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255-1260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 170] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 165. | Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 785] [Cited by in F6Publishing: 742] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 166. | Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126-3130. [PubMed] [Cited in This Article: ] |
| 167. | Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Klöppel G, Kalthoff H. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798-802. [PubMed] [Cited in This Article: ] |
| 168. | Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025-3033. [PubMed] [Cited in This Article: ] |
| 169. | Scarpa A, Capelli P, Mukai K, Zamboni G, Oda T, Iacono C, Hirohashi S. Pancreatic adenocarcinomas frequently show p53 gene mutations. Am J Pathol. 1993;142:1534-1543. [PubMed] [Cited in This Article: ] |
| 170. | Iacobuzio-Donahue CA, Song J, Parmiagiani G, Yeo CJ, Hruban RH, Kern SE. Missense mutations of MADH4: characterization of the mutational hot spot and functional consequences in human tumors. Clin Cancer Res. 2004;10:1597-1604. [PubMed] [Cited in This Article: ] |
| 171. | Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350-353. [PubMed] [Cited in This Article: ] |
| 172. | Goggins M, Offerhaus GJ, Hilgers W, Griffin CA, Shekher M, Tang D, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+. Am J Pathol. 1998;152:1501-1507. [PubMed] [Cited in This Article: ] |
| 173. | Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360-5364. [PubMed] [Cited in This Article: ] |
| 174. | Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, Shekher MC, Westerman AM, Entius MM, Goggins M, Yeo CJ. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154:1835-1840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 270] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 175. | Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329-5332. [PubMed] [Cited in This Article: ] |
| 176. | Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, Hruban RH, Kern SE. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339-2342. [PubMed] [Cited in This Article: ] |
| 177. | Teng DH, Perry WL, Hogan JK, Baumgard M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177-4182. [PubMed] [Cited in This Article: ] |
| 178. | Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, Boehm BO, Pfeifer GP, Hoang-Vu C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806-3812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 122] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 179. | Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, Hidalgo M, Goggins M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146-1156. [PubMed] [Cited in This Article: ] |
| 180. | Thu KL, Radulovich N, Becker-Santos DD, Pikor LA, Pusic A, Lockwood WW, Lam WL, Tsao MS. SOX15 is a candidate tumor suppressor in pancreatic cancer with a potential role in Wnt/β-catenin signaling. Oncogene. 2014;33:279-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 181. | Waraya M, Yamashita K, Katoh H, Ooki A, Kawamata H, Nishimiya H, Nakamura K, Ema A, Watanabe M. Cancer specific promoter CpG Islands hypermethylation of HOP homeobox (HOPX) gene and its potential tumor suppressive role in pancreatic carcinogenesis. BMC Cancer. 2012;12:397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 182. | Chang VH, Chu PY, Peng SL, Mao TL, Shan YS, Hsu CF, Lin CY, Tsai KK, Yu WC, Ch’ang HJ. Krüppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am J Pathol. 2012;181:423-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 183. | Li M, Zhao ZW. Clinical implications of mismatched repair gene promoter methylation in pancreatic cancer. Med Oncol. 2012;29:970-976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 184. | Vogt M, Munding J, Grüner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 248] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 185. | Gao J, Song J, Huang H, Li Z, Du Y, Cao J, Li M, Lv S, Lin H, Gong Y. Methylation of the SPARC gene promoter and its clinical implication in pancreatic cancer. J Exp Clin Cancer Res. 2010;29:28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 186. | Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, Yang D, Omura N, Eshleman J, Canto M. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev. 2008;17:3536-3542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 187. | Bu XM, Zhao CH, Zhang N, Gao F, Lin S, Dai XW. Hypermethylation and aberrant expression of secreted frizzled-related protein genes in pancreatic cancer. World J Gastroenterol. 2008;14:3421-3424. [PubMed] [Cited in This Article: ] |
| 188. | Zhang L, Gao J, Li Z, Gong Y. Neuronal pentraxin II (NPTX2) is frequently down-regulated by promoter hypermethylation in pancreatic cancers. Dig Dis Sci. 2012;57:2608-2614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 189. | Khursheed M, Kolla JN, Kotapalli V, Gupta N, Gowrishankar S, Uppin SG, Sastry RA, Koganti S, Sundaram C, Pollack JR. ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br J Cancer. 2013;108:2056-2062. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 190. | Zhang L, Gao J, Li L, Li Z, Du Y, Gong Y. The neuronal pentraxin II gene (NPTX2) inhibit proliferation and invasion of pancreatic cancer cells in vitro. Mol Biol Rep. 2011;38:4903-4911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 191. | Gu YM, Ma YH, Zhao WG, Chen J. Dickkopf3 overexpression inhibits pancreatic cancer cell growth in vitro. World J Gastroenterol. 2011;17:3810-3817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 192. | Ito T, Kojima T, Yamaguchi H, Kyuno D, Kimura Y, Imamura M, Takasawa A, Murata M, Tanaka S, Hirata K. Transcriptional regulation of claudin-18 via specific protein kinase C signaling pathways and modification of DNA methylation in human pancreatic cancer cells. J Cell Biochem. 2011;112:1761-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 193. | Cai HH, Sun YM, Miao Y, Gao WT, Peng Q, Yao J, Zhao HL. Aberrant methylation frequency of TNFRSF10C promoter in pancreatic cancer cell lines. Hepatobiliary Pancreat Dis Int. 2011;10:95-100. [PubMed] [Cited in This Article: ] |
| 194. | Yao F, Sun M, Dong M, Jing F, Chen B, Xu H, Wang S. NPTX2 hypermethylation in pure pancreatic juice predicts pancreatic neoplasms. Am J Med Sci. 2013;346:175-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 195. | Kato N, Yamamoto H, Adachi Y, Ohashi H, Taniguchi H, Suzuki H, Nakazawa M, Kaneto H, Sasaki S, Imai K. Cancer detection by ubiquitin carboxyl-terminal esterase L1 methylation in pancreatobiliary fluids. World J Gastroenterol. 2013;19:1718-1727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 13] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 196. | Rosty C, Geradts J, Sato N, Wilentz RE, Roberts H, Sohn T, Cameron JL, Yeo CJ, Hruban RH, Goggins M. p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am J Surg Pathol. 2003;27:1495-1501. [PubMed] [Cited in This Article: ] |
| 197. | Hong SM, Omura N, Vincent A, Li A, Knight S, Yu J, Hruban RH, Goggins M. Genome-wide CpG island profiling of intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res. 2012;18:700-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 198. | Park JK, Ryu JK, Lee KH, Lee JK, Yoon WJ, Lee SH, Yoo JW, Woo SM, Lee GY, Lee CH. Quantitative analysis of NPTX2 hypermethylation is a promising molecular diagnostic marker for pancreatic cancer. Pancreas. 2007;35:e9-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 199. | Park JK, Ryu JK, Yoon WJ, Lee SH, Lee GY, Jeong KS, Kim YT, Yoon YB. The role of quantitative NPTX2 hypermethylation as a novel serum diagnostic marker in pancreatic cancer. Pancreas. 2012;41:95-101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 200. | Chen M, Xue X, Wang F, An Y, Tang D, Xu Y, Wang H, Yuan Z, Gao W, Wei J. Expression and promoter methylation analysis of ATP-binding cassette genes in pancreatic cancer. Oncol Rep. 2012;27:265-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 201. | Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136-45.e1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 184] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 202. | Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19:2394-2402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 203. | Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067-1074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 262] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 204. | Zhang S, Cai X, Huang F, Zhong W, Yu Z. Effect of trichostatin a on viability and microRNA expression in human pancreatic cancer cell line BxPC-3. Exp Oncol. 2008;30:265-268. [PubMed] [Cited in This Article: ] |
| 205. | Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38:e190-e199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 216] [Cited by in F6Publishing: 231] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 206. | Park JK, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, Postier RG, Brackett DJ, Schmittgen TD. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406:518-523. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 207. | Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170-16175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 414] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 208. | Mees ST, Mardin WA, Wendel C, Baeumer N, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. EP300--a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int J Cancer. 2010;126:114-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 209. | Hamada S, Satoh K, Miura S, Hirota M, Kanno A, Masamune A, Kikuta K, Kume K, Unno J, Egawa S. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol. 2013;228:1255-1263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 210. | Takikawa T, Masamune A, Hamada S, Nakano E, Yoshida N, Shimosegawa T. miR-210 regulates the interaction between pancreatic cancer cells and stellate cells. Biochem Biophys Res Commun. 2013;437:433-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 211. | Mees ST, Mardin WA, Sielker S, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol. 2009;16:2339-2350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 212. | Chen Z, Chen LY, Dai HY, Wang P, Gao S, Wang K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting Bim expression. J Cell Biochem. 2012;113:3229-3235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 213. | Iwagami Y, Eguchi H, Nagano H, Akita H, Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. 2013;109:502-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 214. | Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 215. | Guo R, Wang Y, Shi WY, Liu B, Hou SQ, Liu L. MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules. 2012;17:14733-14747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 216. | Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, Buscail L, Cordelier P. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831-844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 217. | Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76:5-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 218. | Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 316] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 219. | Li W, Yuan Y, Huang L, Qiao M, Zhang Y. Metformin alters the expression profiles of microRNAs in human pancreatic cancer cells. Diabetes Res Clin Pract. 2012;96:187-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 220. | Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 516] [Cited by in F6Publishing: 522] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 221. | Hu QL, Jiang QY, Jin X, Shen J, Wang K, Li YB, Xu FJ, Tang GP, Li ZH. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. 2013;34:2265-2276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 222. | Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, Ma J, Geng J, Chen Z, Rahman KM. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. 2012;13:1750-1756. [PubMed] [Cited in This Article: ] |
| 223. | Liu C, Cheng H, Shi S, Cui X, Yang J, Chen L, Cen P, Cai X, Lu Y, Wu C. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr Mol Med. 2013;13:467-478. [PubMed] [Cited in This Article: ] |
| 224. | Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 179] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 225. | Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 226. | Pham H, Rodriguez CE, Donald GW, Hertzer KM, Jung XS, Chang HH, Moro A, Reber HA, Hines OJ, Eibl G. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem Biophys Res Commun. 2013;439:6-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 227. | Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. 2012;33:1863-1870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 228. | Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, Maghnouj A, Mirmohammadsadegh A, Hahn SA, Tannapfel A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. 2011;91:1472-1479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 229. | Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther. 2013;12:83-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 230. | Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832-1839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 231. | Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704-6712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 517] [Cited by in F6Publishing: 538] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 232. | Zhou J, Song S, Cen J, Zhu D, Li D, Zhang Z. MicroRNA-375 is downregulated in pancreatic cancer and inhibits cell proliferation in vitro. Oncol Res. 2012;20:197-203. [PubMed] [Cited in This Article: ] |
| 233. | Heyn H, Schreek S, Buurman R, Focken T, Schlegelberger B, Beger C. MicroRNA miR-548d is a superior regulator in pancreatic cancer. Pancreas. 2012;41:218-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 234. | Millward M, Price T, Townsend A, Sweeney C, Spencer A, Sukumaran S, Longenecker A, Lee L, Lay A, Sharma G. Phase 1 clinical trial of the novel proteasome inhibitor marizomib with the histone deacetylase inhibitor vorinostat in patients with melanoma, pancreatic and lung cancer based on in vitro assessments of the combination. Invest New Drugs. 2012;30:2303-2317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 235. | Pili R, Salumbides B, Zhao M, Altiok S, Qian D, Zwiebel J, Carducci MA, Rudek MA. Phase I study of the histone deacetylase inhibitor entinostat in combination with 13-cis retinoic acid in patients with solid tumours. Br J Cancer. 2012;106:77-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 236. | Wang H, Cao Q, Dudek AZ. Phase II study of panobinostat and bortezomib in patients with pancreatic cancer progressing on gemcitabine-based therapy. Anticancer Res. 2012;32:1027-1031. [PubMed] [Cited in This Article: ] |
| 237. | Kanai M, Otsuka Y, Otsuka K, Sato M, Nishimura T, Mori Y, Kawaguchi M, Hatano E, Kodama Y, Matsumoto S. A phase I study investigating the safety and pharmacokinetics of highly bioavailable curcumin (Theracurmin) in cancer patients. Cancer Chemother Pharmacol. 2013;71:1521-1530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 238. | Epelbaum R, Schaffer M, Vizel B, Badmaev V, Bar-Sela G. Curcumin and gemcitabine in patients with advanced pancreatic cancer. Nutr Cancer. 2010;62:1137-1141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 239. | Kanai M, Yoshimura K, Asada M, Imaizumi A, Suzuki C, Matsumoto S, Nishimura T, Mori Y, Masui T, Kawaguchi Y. A phase I/II study of gemcitabine-based chemotherapy plus curcumin for patients with gemcitabine-resistant pancreatic cancer. Cancer Chemother Pharmacol. 2011;68:157-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 283] [Article Influence: 20.2] [Reference Citation Analysis (0)] |