Article Text
Abstract
Objective A prophylactic vaccine is needed to control the HCV epidemic, with genotypes 1–3 causing >80% of worldwide infections. Vaccine development is hampered by HCV heterogeneity, viral escape including protection of conserved neutralising epitopes and suboptimal efficacy of HCV cell culture systems. We developed cell culture-based inactivated genotype 1–3 HCV vaccine candidates to present natively folded envelope proteins to elicit neutralising antibodies.
Design High-yield genotype 1a, 2a and 3a HCV were developed by serial passage of TNcc, J6cc and DBN3acc in Huh7.5 cells and engineering of acquired mutations detected by next-generation sequencing. Neutralising epitope exposure was determined in cell-based neutralisation assays using human monoclonal antibodies AR3A and AR4A, and polyclonal antibody C211. BALB/c mice were immunised with processed and inactivated genotype 1a, 2a or 3a viruses using AddaVax, a homologue of the licenced adjuvant MF-59. Purified mouse and patient serum IgG were assayed for neutralisation capacity; mouse IgG and immune-sera were assayed for E1/E2 binding.
Results Compared with the original viruses, high-yield viruses had up to ~1000 fold increased infectivity titres (peak titres: 6–7 log10 focus-forming units (FFU)/mL) and up to ~2470 fold increased exposure of conserved neutralising epitopes. Vaccine-induced IgG broadly neutralised genotype 1–6 HCV (EC50: 30–193 µg/mL; mean 71 µg/mL), compared favourably with IgG from chronically infected patients, and bound genotype 1–3 E1/E2; immune-sera endpoint titres reached up to 32 000.
Conclusion High-yield genotype 1–3 HCV could be developed as basis for inactivated vaccine candidates inducing broadly neutralising antibodies in mice supporting further preclinical development.
- HCV
- hepatitis C
- chronic viral hepatitis
- molecular biology
- immune response
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
WHAT IS ALREADY KNOWN ON THIS TOPIC
A prophylactic vaccine is required to control the ongoing HCV epidemic.
HCV shows large genetic heterogeneity, with genotypes 1, 2 and 3 causing >80% of infections worldwide.
Neutralising antibodies are key immune correlates for protection in vaccine settings and can protect against chronic HCV infection.
Such protection is associated with antibodies broadly neutralising different HCV genotypes and targeting conserved neutralising epitopes.
For HCV, a whole virus inactivated vaccine strategy is attractive because of its ability to present the HCV envelope glycoprotein complex in its native conformation and to induce neutralising antibodies.
Development of such a vaccine is hampered by low HCV yields in currently available cell culture systems.
WHAT THIS STUDY ADDS
Based on previously developed cell culture infectious HCV recombinants, we developed high-yield genotype 1, 2 and 3 HCV recombinants growing to high titres in cell culture.
Compared with the original viruses, high-yield viruses showed increased exposure of conserved epitopes targeted by neutralising antibodies important for protection against HCV infection.
Vaccine candidates based on inactivated genotype 1, 2 or 3 HCV, respectively, and an adjuvant analogue to the licenced MF-59 adjuvant had the capacity to induce antibodies broadly neutralising HCV of all major genotypes with recognised epidemiological importance.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study is a milestone in the preclinical development of an inactivated HCV vaccine candidate.
Following finalisation of preclinical development, including optimisation of vaccine production conditions, the identified vaccine candidates could enter clinical development.
A prophylactic HCV vaccine is an essential tool to prevent the approximately 1.5 million new infections and 290 000 deaths attributed to this virus every year.
Introduction
Hepatitis C virus (HCV) is a highly prevalent, blood-borne enveloped positive-sense single strand RNA virus of the Flaviviridae family.1 In contrast to the non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B), the structural proteins, capsid protein core and envelope glycoproteins E1 and E2 constitute the viral particle. The E1/E2 heterodimer is the main target for neutralising antibodies (nAb).2 Among eight major genotypes differing in ~30% of their sequence, genotypes 1, 2 and 3 cause >80% of infections worldwide. Genotypes 4, 5 and 6 show a more restricted geographic localisation in the Middle East/Africa, South Africa and Southeast Asia, respectively, while genotypes 7 and 8 were reported in few individuals.1 Subtypes (a, b, c, etc) differ in ~20% of their sequence.
Each year, ~1.5 million new infections occur. Of these, ~80% proceed to chronic infection resulting in a total of ~58 million chronically infected individuals with increased risk for liver cirrhosis and hepatocellular carcinoma, causing ~290 000 deaths annually.3 So far antiviral treatment did not have a major impact on this epidemic, mainly due to lack of symptoms prior to severe liver damage, lack of screening programmes and high treatment costs in many countries.1 3 A prophylactic vaccine will be required to reach the WHO aim to eliminate hepatitis as a major public health threat.
During natural infection, T and B cells appear to contribute to protective immunity. A T cell vaccine using a viral vector approach did not protect against chronic infection in chimpanzees and humans.4 5 In contrast, a B cell vaccine based on E1/E2 glycoprotein heterodimers showed protective effects in chimpanzees and induced nAb in non-human primates, chimpanzees and humans, even though nAb were induced in <50% of human vaccine recipients with limited capacity to neutralise different HCV genotypes.6–10 Induction of nAb correlates with efficacy of other viral vaccines.11 12 Moreover, during natural HCV infections induction of broadly nAb mediated protection.13–15 Protective nAb targeted conserved conformational neutralising epitopes in E2 and E1/E2 localising to antigenic regions 3 and 4 (AR3 and AR4)2 14–16 also targets of well-defined human monoclonal antibodies (mAb).17 18 For efficacy against different HCV genotypes, a future vaccine should target such epitopes which, however, appear to be hidden by closed envelope protein conformational states (E1/E2 states).19 20 Another approach might be a multivalent vaccine based on different viral variants.
For HCV, in mice whole virus vaccines showed a higher capacity to induce nAb than protein-based vaccines,21 presumably due to higher density and more native conformation of the envelope proteins. Indeed, many licenced viral vaccines are based on whole viruses or virus-like particles.12 22 However, application of this technology in HCV vaccine development was hampered by relatively low viral yields in cell culture systems for production of HCV. In 2005, the first systems were developed based on a single genotype 2a isolate (JFH1),23 24 followed by JFH1-based systems expressing genotype specific proteins25 and full-length systems not depending on JFH1.26–28 These systems typically yield 103–105 infectious viruses per mL, considered suboptimal for vaccine development. Nevertheless, proof of concept for immunogenicity of a JFH1-based genotype 2a recombinant was obtained in mice and non-human primates.21 29 However, efficient nAb were only induced with adjuvants not licenced for human use.
Aims of this study were to: (1) develop high-yield culture systems for production of genotype 1a, 2a and 3a HCV, (2) characterise neutralising epitope exposure of high-yield HCV focusing on conserved epitopes associated with protection; and (3) obtain proof of concept for immunogenicity of each high-yield HCV in mice using an adjuvant applicable for human use focusing on detection of antibodies broadly neutralising genotype 1–6 HCV.
Material and methods
Most sections are further detailed in online supplemental materials.
Supplemental material
HCV recombinants
Original TNcc, J6cc and DBN3acc recombinants were developed previously.26–28 High-yield HI-recombinants were engineered using subclones of cell culture derived reverse transcription PCR (RT-PCR) amplicons and In-Fusion technology. Recombinants with genotype(isolate) 1a(TN), 1b(J4), 2a(J6), 2b(J8), 3a(S52), 3a(DBN), 4a(ED43), 5a(SA13) and 6a(HK6a)24 25 30 specific core-NS2 and remaining sequences of genotype 2a isolate JFH1 were used in in vitro HCV neutralisation assays.
Cells
Human hepatoma Huh7.5 cells were used for propagation of HCV. Human embryonic kidney HEK293 cells were used for production of HCV E1/E2 complexes.
Transfection of HCV in vitro RNA transcripts in Huh7.5 cells
Transcripts were produced using T7 RNA polymerase (Promega); transfections were done using Lipofectamine2000 (Invitrogen).25
Infection of Huh7.5 cells with HCV
Cells were inoculated with supernatants derived from transfection experiments at the peak of infection.25
Serial passage of HCV in Huh7.5 cells
Cells were inoculated with culture supernatant derived from the previous passage at the peak of infection.31
Generation of virus stocks in Huh7.5 cells
Cells were inoculated with culture supernatants derived at the peak of infection. Supernatants collected around the peak of infection were pooled. For virus stocks used in neutralisation assays, the HCV envelope protein sequence was confirmed by Sanger sequencing. For virus seed stocks for vaccine virus production, the complete HCV open reading frame (ORF) was analysed by next-generation sequencing (NGS).
Immunostaining of HCV antigens in cell culture
Percentage of HCV infected cells was monitored by immunostaining using primary antibodies monoclonal anti-HCV core antibody C7-50 (EnzoLifeSciences, Farmingdale, New York, USA) diluted 1:5000 and monoclonal anti-HCV NS5A antibody 9E1024 diluted 1:5000 as well as secondary antibody Alexa Fluor 488 goat antimouse IgG (H+L) (Invitrogen) diluted 1:500.25
Determination of HCV infectivity titres
HCV infectivity titres in culture supernatants were determined as focus-forming units (FFU)/mL by titration on 96-well plates and subsequent immunostaining using primary antibodies anti-HCV core antibody C7-50 (EnzoLifeSciences) diluted 1:1000 and anti-HCV NS5A antibody 9E10 diluted 1:3000 as well as secondary antibody ECL sheep antimouse IgG diluted 1:500, followed by visualisation and automated counting of FFU.32
Sequencing of cell culture derived HCV
HCV RNA was extracted from culture supernatants, and either the complete ORF (serial passage experiments, first passage kinetic experiments and virus seed stocks) or E1/E2 (virus stocks for neutralisation assays) were amplified by RT-PCR using specific primers (online supplemental tables 1 and 2) followed by NGS (ORF amplicons) or Sanger sequencing (E1/E2 amplicons).32 33
Subclonal analysis
Selected RT-PCR ORF amplicons were subcloned using the TOPO-XL Cloning kit (Invitrogen) followed by Sanger sequencing.
HCV production for vaccine generation
HCV was produced in serum-free medium in 10-layer cell factories.34 35
Processing of HCV for vaccine generation
HCV was clarified using 5 µm and 0.65 µm filters and concentrated by tangential flow filtration (TFF) with molecular weight cut-off (MWCO) 500 kDa,36 followed by two ultracentrifugation steps using Optiprep Density Gradient Medium (Sigma) for formation of three density cushions and a continuous gradient, respectively, separated by an intermediate TFF step (MWCO 500 kDa). Following size exclusion chromatography using Sephadex G-100 (Sigma Aldrich), HCV was UV irradiated with a UVG-54 Handheld UV lamp (240 nm UV, 6 watt) (Analytik Jena).35
Immunisation of mice
BALB/c mice aged 6–8 weeks (Taconic Farms, Denmark) were subcutaneously immunised four times every 3 weeks with HCV or ovalbumin (OVA) formulated with adjuvant AddaVax 50%/50% (v/v).
Patient samples
Sera or plasma from patients with chronic hepatitis C (CHC) were collected between May 2011 and August 2021 in biobanks attached to the Danish Database for Hepatitis B and C and the HCV Tandem cohort at the Department of Infectious Diseases, Copenhagen University Hospital-Hvidovre. Patients were ≥18 years, had no previous history of treatment for CHC, no coinfection with human immunodeficiency virus (HIV) or hepatitis B virus and no recent intravenous drug use.
Purification, concentration and quantification of IgG
IgG was purified from mouse serum or patient serum or plasma with the Amicon Pro Affinity Concentration Kit Protein G with 50 kDa Amicon Ultra-0.5 Device (Merck Millipore) and concentrated with the Vivaspin 500, 30 000 MWCO (GE Lifescience) kit. Mouse and patient IgG was quantified with the IgG (TOTAL) mouse uncoated ELISA Kit (ThermoFisher) and the Cedex Bio Analyzer (Roche), respectively.
In vitro neutralisation assay
HCV neutralisation with mAb AR3A and AR4A17 18 and polyclonal antibody C21119 was done in a volume of 100 µL followed by inoculation of Huh7.5 cells plated on 96-well plates, subsequently subjected to immunostaining of HCV antigen and automated FFU counting.31 34 HCV neutralisation with purified mouse or patient IgG was done similarly in a volume of 10 µL.37 Percentage of neutralisation was calculated as 100-[100×(FFU count in experimental wells)/(mean FFU count in virus only wells)].
HCV E1/E2 complex ELISA
E1/E2 complexes were obtained from lysates of HEK293 cells transfected with E1/E2 expression plasmids. Binding of mouse IgG or immune-sera to E1/E2 complexes was evaluated by ELISA using secondary antibody ECL sheep antimouse IgG horseradish-peroxidase linked whole antibody (GE Healthcare) diluted 1:1000. Positive controls were mAb AP3338 and H77.39.39 Negative control was secondary antibody only.
Patient and public involvement statement
Patients or the public were not involved in design, conduct, reporting or dissemination plans of our research.
Results
Generation of high-yield genotype 1a, 2a and 3a polyclonal HCV
To develop high-yield culture systems, we serially passaged full-length TNcc (genotype 1a),26 J6cc (genotype 2a)27 and DBN3acc (genotype 3a)28 HCV recombinants in Huh7.5 hepatoma cells until peak HCV infectivity titres showed a plateau at ~6 log10 FFU/mL (figure 1). An additional criterion for termination of passaging was detection of putative cell culture adaptive substitutions in >80% of the viral population as determined by NGS. For an initial TNcc passage line, NGS suggested viral quasispecies populations with mutations at a prevalence <80%, spurring a later passage line with a total of 41 viral passages. For J6cc and DBN3acc, 43 and 22 passages were done, respectively. Compared with the initial passages, late passages showed an increase in HCV infectivity titres of up to 2.6 log10 for TNcc, 1.7 log10 for J6cc and 1.3 log10 for DBN3acc.
In vitro serial passage resulted in high-yield polyclonal genotype 1a, 2a and 3a HCV. Huh7.5 cells were inoculated with cell culture supernatant containing the specified viruses derived from the previous passage culture at the peak of infection, as determined by immunostaining. Every 2–3 days, passage cultures were split, and % HCV antigen positive cells and HCV infectivity titres were determined by immunostaining and infectivity titration, respectively. Peak supernatant HCV infectivity titres determined for the specified passages are means of three replicates with standard deviations (SD). For TNcc, the later passage line was inoculated with passage 10 virus from an initial passage line. Blue frames, passage subjected to NGS. Green frame, passage used for production of genotype 2a seed stock. Red frame, passage subjected to NGS and used for production of genotype 1a seed stock. The genotype 3a seed stock was based on the later developed DBNcc-HI recombinant. FFU, focus-forming unit; NGS, next-generation sequencing.
Genetic changes acquired by high-yield polyclonal genotype 1a, 2a and 3a HCV
To identify genetic correlates of high-yield phenotypes, we carried out NGS of the entire ORF of polyclonal passage (PP) viruses. For TNcc-PP-10 and TNcc-PP-18, derived from passage 10 and 18 of the initial passage line, NGS suggested a viral quasispecies population with most coding nucleotide changes present in <80% of viral genomes (online supplemental table 3). In contrast, for TNcc-PP-38.1 derived from the later passage line, as well as J6cc-PP-35 and DBNcc-PP-16, a more homogeneous viral population was found with most coding nucleotide changes present in ≥80% of viral genomes (figure 2 and online supplemental tables 4–6). TNcc-PP-38.1, J6cc-PP-35 and DBNcc-PP-16 had 17, 17 and 7 coding changes in ≥80% of viral genomes, among which 4, 3 and 1 localised to the envelope proteins, respectively. Of note, DBN3acc already harboured five coding changes in the envelope proteins compared with the consensus HCV sequence in the infected patient this recombinant was based on, while TNcc and J6cc did not contain coding changes in the envelope proteins.26–28 Subclonal analysis of these PP-viruses and phylogenetic analysis of TNcc-PP subclones reflected NGS results (online supplementary results, online supplemental figure 1, online supplemental tables 7–9).
Nucleotide changes in serially passaged genotype 1a, 2a and 3a HCV. For TNcc-PP-38.1, J6cc-PP-35 and DBNcc-PP-16, coding and silent changes with allele frequency >10% determined by NGS of the complete ORF are shown. Coding changes with frequency of ≥80% are specified above bars. Genome positions relate to TNcc,26 J6cc27 and DBN3acc28(GenBank accession numbers JX993348, JQ745650 and KX280714, respectively). For genome positions relating to the H77 (AF009606) reference genome and encoded amino acid changes, see online supplemental tables 4–6. NGS, next-generation sequencing; ORF, open reading frame.
Engineering of high-yield genotype 1a, 2a and 3a HCV recombinants
Based on genetic analysis of high-yield PP-viruses, we engineered high-yield (HI)-recombinants: TNcc-HI-18A and TNcc-HI-18B reflected the two main populations in the initial passage line (online supplemental figure 1 and online supplemental table 3). TNcc-HI reflecting TNcc-PP-38.1 in the later passage line, J6cc-HI reflecting J6cc-PP-35 and DBNcc-HI reflecting DBNcc-PP-16 harboured coding nucleotide changes with >80% frequency in NGS in combinations confirmed by subclonal analysis; as an exception, TNcc-HI also harboured G32S found at 48% frequency (unless otherwise indicated, all amino acid position numbers relate to the H77 reference polyprotein, Genbank accession number AF009606) (figure 2 and online supplemental tables 4–9).
Compared with the original recombinants, all HI-recombinants showed increased fitness in transfection and first passage infection kinetic experiments, with accelerated spread kinetics, monitored by determination of the % infected cells and of HCV infectivity titres and with increased peak infectivity titres. TNcc-HI-18A and TNcc-HI-18B peak infectivity titres were only approaching 5 log10 FFU/mL and thus fell short of the target of 6 log10 FFU/mL (online supplemental figure 2). In contrast, in transfection/infection experiments, TNcc-HI, J6cc-HI and DBNcc-HI yielded peak infectivity titres of 5.8/6.0, 6.1/6.8 and 6.4/7.0 log10 FFU/mL, respectively, while the respective original recombinants yielded 3.0/3.5, 3.5/3.4 and 5.3/5.3 log10 FFU/mL, respectively (figure 3). In infection experiments, infectivity titres of HI-viruses were comparable with those of PP-viruses (figure 3B). HI-recombinants were genetically stable following first viral passage (no acquisition of substitutions with >10% frequency with exception of TNcc-HI that acquired L179P and S1930Y with 15% and 39% frequency, respectively).
Engineered high-yield genotype 1a, 2a and 3a HCV recombinants had increased viral fitness in cell culture. (A) Specified recombinants were transfected into Huh7.5 cells using the same amount of HCV RNA in vitro transcripts for recombinants directly compared in each graph. (B) In first passage kinetic experiments, cells were infected at multiplicity of infection of 0.002 with original and HI-viruses using supernatants derived from the transfection experiment when peak infectivity titres were observed; polyclonal passage viruses were included for comparison. (A and B) Every 2–3 days, passage cultures were split, and % HCV antigen positive cells and HCV infectivity titres were determined by immunostaining and infectivity titration, respectively. HCV infectivity titres are means of three replicates with SD. FFU, focus-forming unit; MOI, multiplicity of infection.
High-yield genotype 1a, 2a and 3a recombinants showed increased exposure of neutralising epitopes
Compared with the respective recombinants with in vivo derived envelope protein sequences without cell culture adaptive substitutions,24 30 based on determined EC50 values, HI-recombinants showed 12-fold to 2472-fold increased sensitivity to neutralisation by human-derived mAb AR3A17 and AR4A,18 targeting conserved conformational epitopes in E2 and E1/E2 associated with protection, respectively, and by polyclonal IgG C21119 derived from a patient chronically infected with genotype 1a (figure 4). In detail, TNcc-HI showed 300-fold, 2400-fold and 110-fold increased neutralisation sensitivity to AR3A, AR4A and C211, respectively, while a genotype 1a HCV seed stock derived from TN-PP-18 showed 3-fold, 15-fold and 3.2-fold increased sensitivity. J6cc-HI showed 440-fold, 12-fold and 633-fold increased neutralisation sensitivity. DBNcc-HI showed 1250-fold, 220-fold and 2472-fold, while DBN3acc showed 167-fold, 22-fold and 88-fold increased neutralisation sensitivity. For viruses with in vivo derived TN, J6 and DBN envelope protein sequences, determined half maximal effective concentrations (EC50) were in line with previous results.20 40 41 Thus, HI-viruses showed greatly increased exposure of conserved conformational neutralising epitopes associated with protection against chronic HCV infection.
Engineered high-yield genotype 1a, 2a and 3a HCV recombinants showed increased sensitivity to neutralisation by human-derived nAb. Recombinants with in vivo derived genotype(isolate) 1a(TN),30 2a(J6)24 and 3a(DBN)30 core-NS2 sequences, as well as TNcc-HI, J6cc-HI, DBNcc-HI, the genotype 1a HCV seed stock and DBN3acc28 with envelope protein substitutions acquired during in vitro passage were subjected to neutralisation with human mAb AR3A and AR4A,17 18 and polyclonal antibody C211.19 Data points are means of three replicates with SD; curves were fitted, and EC50 were calculated with the formula y=100/(1+10(log10EC50−X)×hillslope) using GraphPad prism. Fold increase in neutralisation sensitivity was calculated as [(EC50 of 1a(TN), 2a(J6) or 3a(DBN) virus with in vivo derived envelope protein sequence)/(EC50 of respective virus with in vitro derived envelope protein substitutions)]. Virus stock envelope protein sequences were confirmed by Sanger sequencing. EC50, half maximal effective concentration; gt, genotype; mAb, monoclonal antibody; nAb, neutralising antibody.
Generation of vaccine candidates based on inactivated high-yield genotype 1a, 2a and 3a recombinants
To produce viruses for vaccine experiments, HCV seed stocks were generated by inoculation of Huh7.5 cells with polyclonal virus preparation TNcc-PP-18, polyclonal virus preparation J6cc-PP-38 or a first viral passage DBNcc-HI virus, available on initiation of vaccine studies. Sequence confirmed genotype 1a, 2a and 3a HCV seed stocks with infectivity titres of 4.8, 6.2 and 6.4 log10 FFU/mL, respectively (online supplemental tables 3, 5 and 6), were used to inoculate Huh7.5 cells for HCV production in 10-layer cell factories, resulting in a total volume of 16 L HCV containing supernatant per virus (online supplemental figure 3). Supernatants were subjected to downstream processing, involving an initial filter clarification followed by two TFF steps, cushion ultracentrifugation, another TFF, gradient ultracentrifugation, chromatography and inactivation by UV irradiation (supplementary results in online supplemental file 1 and online supplemental figures 4 and 5).
Immunisation of mice with inactivated genotype 1a, 2a and 3a HCV vaccine candidates elicited broadly neutralising and envelope-protein binding antibodies
Processed inactivated genotype 1a, 2a or 3a HCVcc or control antigen OVA were formulated with the adjuvant AddaVax, an analogue of the adjuvant MF-59, which is licenced for human use, and used for immunisation of BALB/c mice.
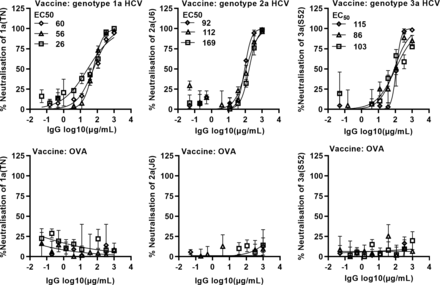
Purified serum IgG from individual animals neutralised HCV with in vivo derived envelope protein sequences of the same genotype as used in the respective vaccine in concentration dependent manner, with mean EC50 of 47, 124 and 101 µg/mL for genotype 1a, 2a and 3a HCV vaccines, respectively (figure 5). Close to complete neutralisation was achieved at the highest applied IgG concentration of 1000 µg/mL. OVA immunised mice did not elicit HCV nAb.
Immunisation with inactivated genotype 1a, 2a or 3a HCV elicited antibodies neutralising cell culture infectious HCV of the same genotype. Groups of three mice were immunised with inactivated genotype 1a, 2a and 3a HCV or OVA formulated with adjuvant AddaVax. Purified serum IgG from individual mice was used to neutralise recombinants containing in vivo derived genotype(isolate) 1a(TN),30 2a(J6)24 and 3a(S52)25 specific core-NS2. Data points are means of three replicates with SD; curves were fitted, and EC50 were calculated with the formula y=100/(1+10(log10EC50−X)×hillslope) using GraphPad prism. Each concentration–response curve specified by unique symbols represents data from one animal. Virus stock envelope protein sequences were confirmed by Sanger sequencing. EC50, half maximal effective concentration; nAb, neutralising antibody; OVA, ovalbumin.
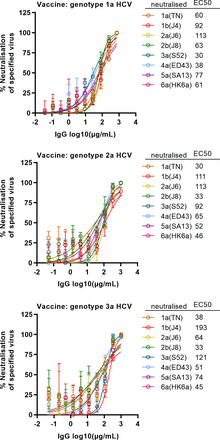
Moreover, IgG pools neutralised genotype 1–5 HCV with in vivo derived envelope sequences and genotype 6 HCV with two vital cell culture adaptive substitutions in the envelope proteins with similar efficacy and in concentration dependent manner. Across neutralised viruses, mean EC50 of IgG from genotype 1a, 2a and 3a vaccinated animals were 67, 68 and 77 µg/mL, respectively, and close to complete neutralisation was observed at 1000 µg/mL (figure 6). Mouse IgG neutralisation capacity compared favourably with that of IgG from patients with CHC regarding efficacy and broadness (figure 7). Interestingly, these data confirmed that 5a(SA13) had relatively high neutralisation sensitivity, while 2a(J6) and 3a(S52) had relatively low neutralisation sensitivity.42 Furthermore, neutralisation capacity of IgG from genotype 3a infected patients was lower than that from genotype 1a, 2a or 2b infected patients.
Immunisation with inactivated genotype 1a, 2a or 3a HCV elicited broadly nAb. Purified IgG from mice of each group, immunised with either genotype 1a, 2a or 3a HCV, was pooled using the same amount of IgG from each animal. IgG pools were used to neutralise recombinants containing in vivo derived genotype(isolate) 1a(TN), 1b(J4), 2a(J6), 2b(J8), 3a(S52), 4a(ED43) and 5a(SA13) core-NS2 sequences; 6a(HK6a) contained two vital cell culture adaptive substitutions in E1 and E2.24 25 30 Data points are means of three replicates with SD; curves were fitted, and EC50 were calculated with the formula y=100/(1+10(log10EC50−X)×hillslope) using GraphPad prism. Virus stock envelope protein sequences were confirmed by Sanger sequencing. EC50, half maximal effective concentration; nAb, neutralising antibody.
Vaccine-induced nAb responses compare favourably with those in patients with chronic HCV infection. Purified IgG from patients chronically infected with HCV of genotype 1a, 2a/2b or 3a at the specified concentrations was used to neutralise recombinants containing in vivo derived genotype(isolate) 1a(TN), 2a(J6), 3a(S52) and 5a(SA13) core-NS2 sequences.24 25 30 Genotype 2a versus 2b patients are indicated by grey versus black open circles. Data points obtained in neutralisation assays with the same concentrations of purified mouse IgG shown in figure 6 are replotted for comparison. All data points are means of three replicates. Virus stock envelope protein sequences were confirmed by Sanger sequencing. gt, genotype; nAb, neutralising antibody.
Immunisation with inactivated genotype 1a, 2a or 3a HCV elicited antibodies binding to HCV envelope proteins. Binding capacity of pooled (A) purified serum IgG or (B and C) immune-sera from mice immunised with inactivated genotype 1a, 2a or 3a HCV or OVA to E1/E2 complexes of the specified HI-recombinants was evaluated by ELISA. Values are optical density (OD) reads at 450 nm following subtraction of mean OD of eight negative controls. Data points are means of two replicates with SD. (A) Positive controls: instead of serum IgG, well-characterised primary antibodies were used: AP3338 for binding to TNcc-HI and DBNcc-HI E1/E2 and H77.3939 for binding to J6cc-HI E1/E2. Negative controls: no IgG or immune-sera were used, and TNcc-HI, J6cc-HI and DBNcc-HI E1/E2 were incubated with secondary antibody only; for negative controls, OD reads were ~0.05. (A and B) In the OVA graphs, data points reflecting binding to J6cc-HI E1/E2 and DBNcc-HI E1/E2 were nudged by 0.04 and 0.08 units in the y direction, respectively. (C) Immune-sera endpoint titres were determined as the highest serum dilution yielding an OD >2 fold mean OD of negative controls. OVA, ovalbumin.
Finally, IgG pools from all HCV immunised animal groups efficiently bound TNcc-HI, J6cc-HI and DBNcc-HI E1/E2 complexes in concentration-dependent manner (figure 8, online supplemental figure 6). Such binding was not observed for pooled IgG from OVA immunised animals. Pools of immune-sera had endpoint titres of up to 32 000 (figure 8, online supplemental figure 7).
Discussion
In this study, we developed high-yield genotype 1a, 2a and 3a HCV cell culture systems to facilitate development of whole virus inactivated vaccine candidates. Compared with the original viruses, high-yield viruses showed increased exposure of conserved conformational neutralising epitopes associated with protection against chronic HCV infection, as suggested by increased sensitivity to neutralisation by mAb AR3A and AR4A. In mouse immunogenicity studies, high-yield genotype 1a, 2a or 3a viruses, formulated with an analogue of the human MF-59 adjuvant, each had the capacity to induce efficient nAb broadly neutralising HCV of all major genotypes with recognised epidemiological importance.
For efficient production of whole virus inactivated vaccines, high-yield virus production is required. For example, SARS-CoV-2 used for vaccine production grow to infectivity titres of 6.5–7 log10 TCID50/mL.43 For selection of high-yield variants of previously developed full-length recombinants of the most prevalent HCV genotypes,26–28 we employed a serial passage approach, previously used for further adaption of a JFH1-based genotype 5a virus.31 Applying this evolutionary approach until no further increase in viral infectivity titres is observed, and until a homogeneous viral population with no obvious evidence for ongoing selection of additional putative adaptive substitutions is recorded is expected to result in selection of highly fit and genetically stable virus populations.31 Thus, the developed genetically stable, high-yield HCV recombinants can in the future be used to initiate virus vaccine antigen production with sequence confirmed early viral passage seed stocks.44 45 Based on results from this study using full-length recombinants, as well as on results from the previous study using a JFH1-based recombinant,31 the upper limit for HCV infectivity titres in monolayer Huh7.5 cell cultures is between 6 and 7 log10 FFU/mL, which might be due to limited availability of required host cell factors. Future studies should focus on investigation of the effect of acquired viral substitutions on the viral life cycle. Interestingly, several substitutions selected in this study were also selected during cell culture adaptation of other HCV recombinants, suggesting a general role for HCV cell culture adaptation (online supplemental table 10).
In future studies, it will be of special interest to investigate which of the selected envelope substitutions conferred increased exposure of the conserved conformational epitopes targeted by AR3A and AR4A. Several genetic changes in E2 such as deletion of HVR1 (aa 384-410),40 abrogation of N-linked glycosylation,19 46 but also specific substitutions in HVR1 (aa 400-404) and the E2 front layer (aa 414, 431 and 453; front layer: aa 411–461)20 were described to increase neutralisation sensitivity, which was linked to an open E1/E2 state.19 20 TN-PP-18 and TN-PP-38.1 acquired N410K in HVR1, while TN-PP-38.1 in addition acquired F403L in HVR1. J6cc-PP-35 acquired H434N in the E2 front layer and DBNcc-PP-16 acquired G395R in HVR1, while DBN3acc harboured S449A in the E2 front layer.28 Furthermore, localising to E2 outside these specific regions previously associated with changes in neutralisation sensitivity, TN-PP-38.1 acquired V719I and J6cc-PP-35 acquired A573T, while DBN3acc harboured D474A, T528N and V629A.28
A positive correlation between viral fitness and neutralising epitope exposure was also observed in previous studies for HCV31 and HIV.47 For HCV, in vivo protection of conserved conformational neutralising epitopes might be associated with a fitness cost as closed E1/E2 states might decrease access of the main HCV entry receptor CD81 to its binding site, which is overlapping with AR3.2
For HCV, deletion of HVR1 led to a maximally open E1/E2 state associated with high neutralisation sensitivity.19 40 For the full length genotype 1–3 HI-viruses developed in this study and the previously developed JFH1-based high-yield genotype 5a HCV,31 the AR3A epitopes were as accessible as in HVR1-deleted viruses, while the AR4A epitopes were approximately 10-fold to 100-fold less exposed.35 40 However, HVR1-deleted viruses typically show relatively low infectivity titres, hampering vaccine production.48 Compared with the original viruses, HI-viruses showed ~300 to 1250-fold higher exposure of AR3A epitopes and ~12 to 2400-fold higher exposure of AR4A epitopes; the high-yield genotype 5a HCV showed only ~10 fold higher exposure of AR3A and AR4A epitopes than the original already highly neutralisation sensitive genotype 5a HCV.35
Therefore, the genotype 1a, 2a and 3a HI-viruses developed in this study and the high-yield JFH1-based genotype 5a HCV31 present interesting vaccine antigens as they might facilitate induction of antibodies targeting epitopes that are conserved between HCV variants and that are mediating protection in humans. A vaccine antigen exposing such conserved epitopes with the ability to induce broadly nAb might make a multivalent vaccine approach unnecessary.
Indeed, immunisation with genotype 1–3 PP-viruses and HI-viruses and with high-yield genotype 5a HCV31 resulted in induction of broadly nAb. Fifty per cent neutralisation titres and ELISA endpoint titres of vaccine-induced antibodies were comparable with those reported for licenced antiviral vaccines and with those in chimpanzees protected from HCV challenge following vaccination with the E1/E2 heterodimer vaccine.6 10 However, compared with IgG from chronically infected patients, antibodies elicited by the E1/E2 heterodimer vaccine8–10 and different vaccine candidates based on soluble E2 protein,46 49–51 nAb induced in this and the previous study35 showed increased capacity to neutralise different HCV variants. Finally, 50% neutralisation titres were comparable with those reported for a genotype 2a inactivated vaccine candidate in mice and non-human primates when experimental adjuvants not suitable for human use were applied.21 29 In the study of non-human primates, application of the licenced adjuvant aluminium hydroxide did not result in induction of efficient nAb.29 AddaVax/MF-59 appears to be more immunogenic than aluminium hydroxide.35 52 Furthermore, for other viruses, increased neutralising epitope exposure was suggested to result in increased immunogenicity.53 54 In future studies, it would be interesting to investigate, whether increased exposure of conserved neutralising epitopes increases immunogenicity and whether different HCV genotypes/serotypes differ in immunogenicity.42 55 However, this would require development of high-yield viruses without E2 substitutions mediating epitope exposure, which might not be possible, as it is likely that increased fitness was at least partly mediated by such substitutions. Soluble E2 or E1/E2 heterodimer vaccine platforms might be more amenable for such studies; however, they might not reflect the native envelope protein conformation on the whole virus particle. In such studies, deletion of the three variable E2 regions was reported to result in a certain increase in immunogenicity, whereas deletion of HVR1 and/or modification of glycosylation sites had no or a minor effect on immunogenicity.46 49–51 56 Deletion of HVR1 of the already highly neutralisation sensitive high-yield genotype 5a HCV, facilitated by subsequent culture adaptation, did not result in increased immunogenicity.35 In addition, future studies requiring larger amounts of HI-virus vaccine induced nAb and most likely derived mAb could investigate which epitopes are targeted by these antibodies.
Further preclinical and clinical development requires optimisation of vaccine production and processing conditions to ensure compatibility with vaccine manufacturing.36 44 Moreover, further research should define the most powerful of the developed vaccine candidates, based on performance in an optimised bioprocess and detailed immunogenicity studies of resulting antigens, as well as dose finding studies. In initial upstream bioprocess studies employing a scalable bioreactor44 57 and virus seed stocks generated from early viral passages following transfection, genotype 2a and 3a HI-viruses and high-yield genotype 5a HCV35 yielded considerably higher infectivity titres than the genotype 1a HI-virus, signifying an advantage for these three candidates in the production process. No immunocompetent HCV in vivo challenge model is available. While future studies might employ specialised small animal models such as the human liver chimeric uPA-SCID mouse model to study certain aspects of vaccine-induced protection48 and larger animals to confirm vaccine safety and immunogenicity,9 promising vaccine candidates likely need to proceed to clinical trials involving controlled human infection models58 to evaluate their true protective potential. Finally, in the quest for an HCV vaccine, it will be important to facilitate cross-comparison of vaccine candidates by application of standardised assays.42 55
In conclusion, we developed high-yield genotype 1a, 2a and 3a HCV constituting a basis for inactivated vaccine candidates that could be used for further preclinical and clinical development.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Ethics approval
The HCV Tandem cohort was approved by the Regional Ethical Committee (H-21004361) and the Danish Data Protection Agency (2012-58-0004). Participants gave informed consent to participate in the study before taking part.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Contributors Study design: GPA, AFP, JB and JMG. Acquisition of data: GPA, AFP, AO, CRDH, ZD, SF and UF. Analysis and interpretation of data: GPA, AFP, AO, CRDH, UF, CS, NW, ML, JCP, JB and JMG. Contribution of materials: CS, NW and ML. Drafting of the manuscript: GPA and JMG. Study supervision: JMG. Guarantor: JMG. All authors reviewed the manuscript.
Funding This work was supported by PhD stipends and bonuses from the Candys Foundation and the University of Copenhagen (AFP, AO, JB and JMG), the China Scholarship Council (ZD and JMG), grants from the Novo Nordisk Foundation (NW, JB and JMG), The Danish Cancer Society (JB and JMG), Independent Research Fund Denmark (DFF) - Medical Sciences (JB and JMG), Innovation Fund Denmark (JB and JMG), The Lundbeck Foundation (JP and JB), The Region H Foundation (AFP, CS, JB and JMG), The Toyota Foundation (AO and JMG), The Læge Sofus Carl Emil Friis og Hustru Olga Doris Friis’ Foundation (JMG) and The Mauritzen La Fontaine Foundation (JB). ML is partly funded by NIH grants AI123861 and AI144232.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.